si:dkey-104n9.1
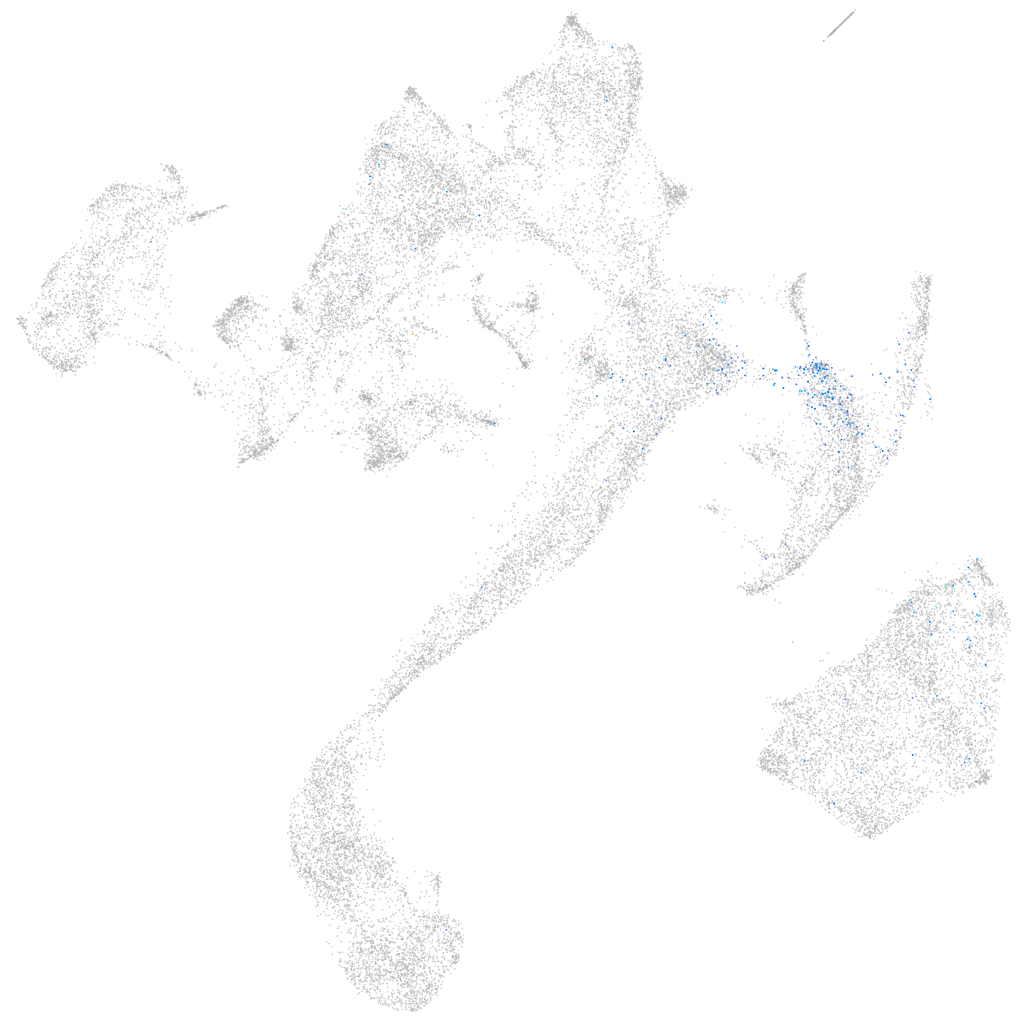
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mespba | 0.205 | fabp3 | -0.062 |
| tbx6 | 0.190 | actc1b | -0.058 |
| pcdh8 | 0.183 | pabpc4 | -0.054 |
| ripply2 | 0.183 | ak1 | -0.053 |
| mespab | 0.171 | bhmt | -0.052 |
| draxin | 0.147 | eno1a | -0.052 |
| ripply1 | 0.146 | ckmb | -0.051 |
| grin2da | 0.139 | ckma | -0.051 |
| her11 | 0.135 | aldoab | -0.051 |
| kazald2 | 0.135 | atp2a1 | -0.050 |
| mespbb | 0.128 | eno3 | -0.049 |
| mespaa | 0.127 | tmem38a | -0.048 |
| cpa6 | 0.124 | gapdh | -0.048 |
| meox1 | 0.124 | mylpfa | -0.047 |
| dlc | 0.122 | tnnc2 | -0.047 |
| zgc:174945 | 0.118 | si:dkey-16p21.8 | -0.047 |
| tjp2b | 0.118 | tpi1b | -0.046 |
| dmrt2a | 0.117 | acta1b | -0.046 |
| tcf15 | 0.116 | sparc | -0.045 |
| adgrl2a | 0.111 | eef1da | -0.045 |
| fn1b | 0.111 | CABZ01078594.1 | -0.045 |
| myf5 | 0.109 | pgk1 | -0.045 |
| lmo4b | 0.109 | plecb | -0.045 |
| large2 | 0.109 | ndrg2 | -0.045 |
| tsc22d2 | 0.104 | pmp22a | -0.044 |
| arl4aa | 0.103 | neb | -0.044 |
| zic3 | 0.103 | srl | -0.044 |
| dld | 0.101 | glud1b | -0.044 |
| gnaia | 0.100 | gamt | -0.044 |
| znfl1k | 0.098 | ssuh2rs1 | -0.044 |
| rasgef1ba | 0.097 | myl1 | -0.043 |
| her1 | 0.097 | atp1a2a | -0.043 |
| apoc1 | 0.096 | pvalb1 | -0.043 |
| tuba8l2 | 0.094 | ldb3a | -0.043 |
| rgs12a | 0.092 | mylz3 | -0.043 |