si:dkey-102g19.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm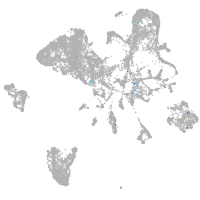 |
axial mesoderm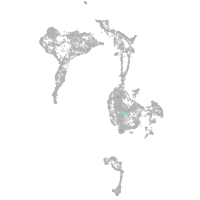 |
muscle 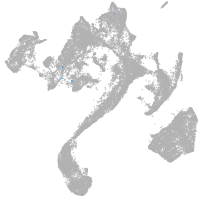 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 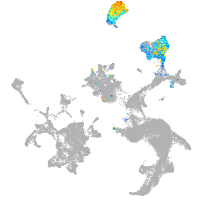 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sc:d156 | 0.755 | mt-atp6 | -0.078 |
| CU499336.2 | 0.746 | mt-co1 | -0.077 |
| CABZ01001434.1 | 0.743 | rpl3 | -0.073 |
| dok2 | 0.732 | ubb | -0.071 |
| itln1 | 0.732 | mt-cyb | -0.070 |
| FERMT3 (1 of many) | 0.727 | ndufb9 | -0.067 |
| CABZ01050944.1 | 0.718 | mt-nd1 | -0.063 |
| LOC101884267 | 0.698 | rpl27a | -0.061 |
| vaspa | 0.665 | ndufb10 | -0.061 |
| tnfaip8l2a | 0.622 | ppdpfb | -0.061 |
| ponzr6 | 0.610 | gdi2 | -0.059 |
| npsn | 0.568 | mt-co2 | -0.059 |
| glipr1a | 0.544 | ndufb7 | -0.058 |
| pdcb | 0.539 | nop53 | -0.057 |
| spi1b | 0.523 | tomm5 | -0.055 |
| rasal3 | 0.501 | ywhaqb | -0.055 |
| wasb | 0.499 | mgst3b | -0.055 |
| CU571069.1 | 0.470 | atp1a1a.1 | -0.055 |
| grap2b | 0.445 | ube2v1 | -0.054 |
| si:dkey-27h10.2 | 0.430 | rps27.2 | -0.054 |
| spice1 | 0.428 | calm2a | -0.054 |
| ecm1a | 0.387 | ndufa4l | -0.053 |
| fcer1gl | 0.386 | paip2b | -0.052 |
| pcolceb | 0.381 | si:ch1073-325m22.2 | -0.050 |
| coro1a | 0.380 | ywhae1 | -0.050 |
| zgc:91999 | 0.373 | ndufab1a | -0.049 |
| trpm3 | 0.368 | ap2m1a | -0.049 |
| lyz | 0.366 | rnaseka | -0.048 |
| AL953886.1 | 0.340 | si:dkey-16p21.8 | -0.048 |
| si:dkey-93h22.4 | 0.333 | psmb7 | -0.048 |
| wipf1b | 0.327 | pnrc2 | -0.048 |
| zgc:101715 | 0.308 | ndufa10 | -0.048 |
| lhcgr | 0.304 | hdlbpa | -0.048 |
| LOC110437795 | 0.295 | ndufs4 | -0.048 |
| sept4 | 0.278 | vamp3 | -0.047 |




