si:dkey-102g19.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm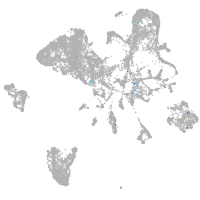 |
axial mesoderm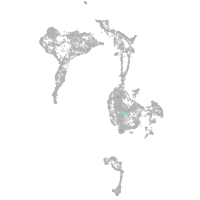 |
muscle 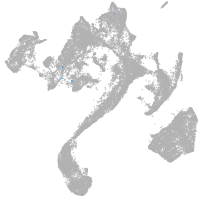 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 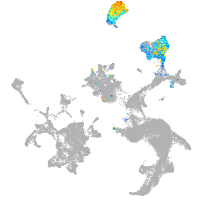 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lect2l | 0.780 | hbae1.1 | -0.286 |
| mmp13a | 0.765 | hbae3 | -0.272 |
| glipr1a | 0.757 | hbbe1.3 | -0.272 |
| npsn | 0.748 | hbae1.3 | -0.271 |
| cfbl | 0.740 | hbbe2 | -0.267 |
| fcer1gl | 0.735 | hbbe1.1 | -0.258 |
| lta4h | 0.730 | cahz | -0.247 |
| mpx | 0.726 | lmo2 | -0.233 |
| ponzr6 | 0.708 | hbbe1.2 | -0.233 |
| alox5ap | 0.707 | hemgn | -0.231 |
| lyz | 0.694 | fth1a | -0.229 |
| spi1b | 0.688 | blvrb | -0.222 |
| il1b | 0.683 | zgc:56095 | -0.220 |
| ctss2.1 | 0.675 | alas2 | -0.217 |
| scpp8 | 0.674 | si:ch211-250g4.3 | -0.215 |
| scinlb | 0.670 | creg1 | -0.210 |
| dhrs9 | 0.663 | slc4a1a | -0.205 |
| cpa5 | 0.650 | nt5c2l1 | -0.200 |
| si:ch211-276a23.5 | 0.650 | epb41b | -0.191 |
| spice1 | 0.649 | zgc:163057 | -0.191 |
| ctss1 | 0.646 | si:ch211-207c6.2 | -0.183 |
| coro1a | 0.644 | nmt1b | -0.175 |
| nccrp1 | 0.643 | aqp1a.1 | -0.174 |
| si:ch211-284o19.8 | 0.642 | prdx2 | -0.169 |
| il34 | 0.640 | tmod4 | -0.168 |
| ch25hl2 | 0.638 | plac8l1 | -0.167 |
| CABZ01001434.1 | 0.632 | arl4aa | -0.158 |
| gsto2 | 0.628 | si:ch211-227m13.1 | -0.156 |
| si:ch211-193e13.5 | 0.624 | sptb | -0.155 |
| si:ch1073-443f11.2 | 0.623 | hbae5 | -0.155 |
| gapdh | 0.621 | uros | -0.155 |
| arpc1b | 0.620 | znfl2a | -0.155 |
| aldh8a1 | 0.615 | tfr1a | -0.152 |
| CR848841.1 | 0.614 | rfesd | -0.147 |
| LOC101884267 | 0.611 | hdr | -0.146 |




