si:dkey-102g19.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 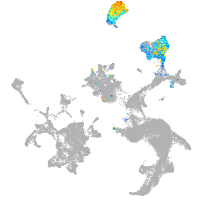 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cga | 0.525 | bace2 | -0.025 |
| h2af1al | 0.451 | LOC103910009 | -0.023 |
| atg4db | 0.368 | ywhaz | -0.023 |
| klf17 | 0.348 | si:ch211-286b5.5 | -0.022 |
| rell2 | 0.348 | pnp4a | -0.022 |
| il10ra | 0.327 | tspan10 | -0.022 |
| hif1al2 | 0.309 | zgc:56493 | -0.021 |
| si:dkeyp-68b7.12 | 0.287 | prdx6 | -0.021 |
| CR391915.1 | 0.285 | coro1ca | -0.021 |
| tec | 0.277 | slc3a2a | -0.020 |
| sowahd | 0.269 | calm2b | -0.020 |
| CU076099.1 | 0.240 | syngr1a | -0.020 |
| XLOC-008693 | 0.236 | zgc:110591 | -0.020 |
| slc6a22.2 | 0.231 | tmem130 | -0.020 |
| pcdh15b | 0.227 | LEPROTL1 | -0.019 |
| traf5 | 0.223 | atp6ap2 | -0.019 |
| gp1bb | 0.219 | calm2a | -0.019 |
| BX649485.2 | 0.209 | ndufa4l | -0.019 |
| LO017816.1 | 0.202 | sigmar1 | -0.018 |
| igfbp5a | 0.198 | aldh6a1 | -0.018 |
| vaspa | 0.194 | cst14b.1 | -0.018 |
| plac8l1 | 0.192 | psenen | -0.018 |
| vstm4b | 0.188 | syngr2b | -0.018 |
| plaua | 0.187 | fabp3 | -0.018 |
| nt5e | 0.185 | bco1 | -0.018 |
| fcer1gl | 0.184 | eif3ha | -0.018 |
| parm1 | 0.182 | ndufb10 | -0.018 |
| si:dkey-242g16.2 | 0.181 | pim1 | -0.018 |
| RF00030 | 0.175 | pmp22b | -0.018 |
| si:dkey-5n18.1 | 0.173 | si:dkey-21a6.5 | -0.018 |
| BX950188.1 | 0.169 | sept8a | -0.018 |
| zgc:172075 | 0.168 | rabl6b | -0.018 |
| sned1 | 0.167 | cdh1 | -0.017 |
| lect2l | 0.166 | atp6v1ab | -0.017 |
| BX323060.1 | 0.163 | cdk15 | -0.017 |