si:ch73-334d15.4
ZFIN 
Other cell groups


all cells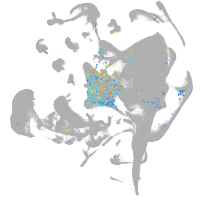 |
blastomeres |
PGCs |
endoderm |
axial mesoderm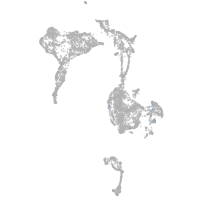 |
muscle 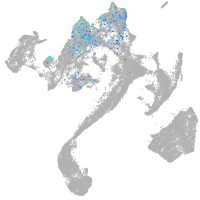 |
mural cells / non-skeletal muscle  |
mesenchyme 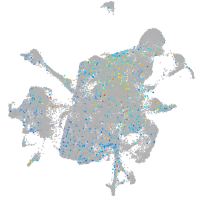 |
hematopoietic / vasculature 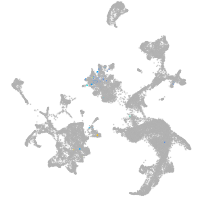 |
pronephros  |
neural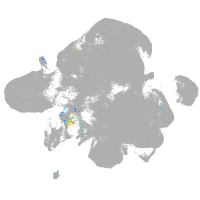 |
spinal cord / glia |
eye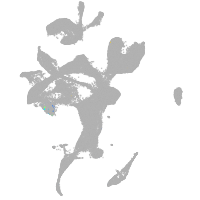 |
taste / olfactory |
otic / lateral line |
epidermis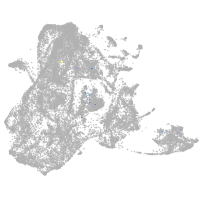 |
periderm |
fin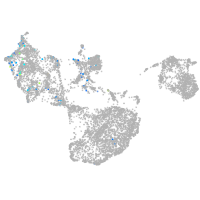 |
ionocytes / mucous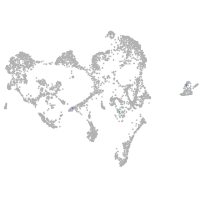 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dnase1l1l | 0.517 | syngr1a | -0.030 |
| mespba | 0.517 | qdpra | -0.027 |
| zgc:173915 | 0.517 | rab38 | -0.027 |
| pgfb | 0.496 | tspan36 | -0.026 |
| BX248410.3 | 0.435 | ywhaz | -0.025 |
| BX511128.3 | 0.435 | gpr143 | -0.025 |
| col8a1b | 0.435 | rabl6b | -0.025 |
| CR382296.1 | 0.412 | rab34b | -0.024 |
| ACOT12 | 0.406 | tmem243a | -0.024 |
| si:dkey-208k4.3 | 0.405 | C12orf75 | -0.023 |
| LOC110438943 | 0.382 | smim29 | -0.023 |
| tbx22 | 0.352 | cfl1 | -0.023 |
| glt8d2 | 0.339 | arhgdia | -0.022 |
| ccl38.1 | 0.322 | elovl1b | -0.022 |
| rsad2 | 0.321 | sypl2b | -0.022 |
| rbp7b | 0.297 | marcksl1b | -0.022 |
| slc6a22.2 | 0.294 | slc2a15a | -0.021 |
| pappab | 0.285 | cd164 | -0.021 |
| LOC103909982 | 0.283 | pmela | -0.021 |
| CR932000.1 | 0.278 | rab1ab | -0.021 |
| loxl5a | 0.277 | pcbd1 | -0.021 |
| lrrc15 | 0.275 | prdx1 | -0.020 |
| LOC101882264 | 0.275 | opn5 | -0.020 |
| CR391910.2 | 0.274 | mtbl | -0.020 |
| tmem173 | 0.268 | gstt1a | -0.020 |
| LOC110439889 | 0.262 | idh1 | -0.020 |
| nog3 | 0.252 | rab27a | -0.020 |
| slc6a4a | 0.250 | dct | -0.019 |
| CR388231.2 | 0.243 | cotl1 | -0.019 |
| trpc6a | 0.242 | tyrp1b | -0.019 |
| nanos2 | 0.228 | palm1b | -0.019 |
| gata1a | 0.228 | tyr | -0.019 |
| c1qtnf2 | 0.226 | degs1 | -0.019 |
| col27a1a | 0.226 | tuba8l3 | -0.019 |
| serpinf1 | 0.224 | pah | -0.019 |




