si:ch73-122g19.1
ZFIN 
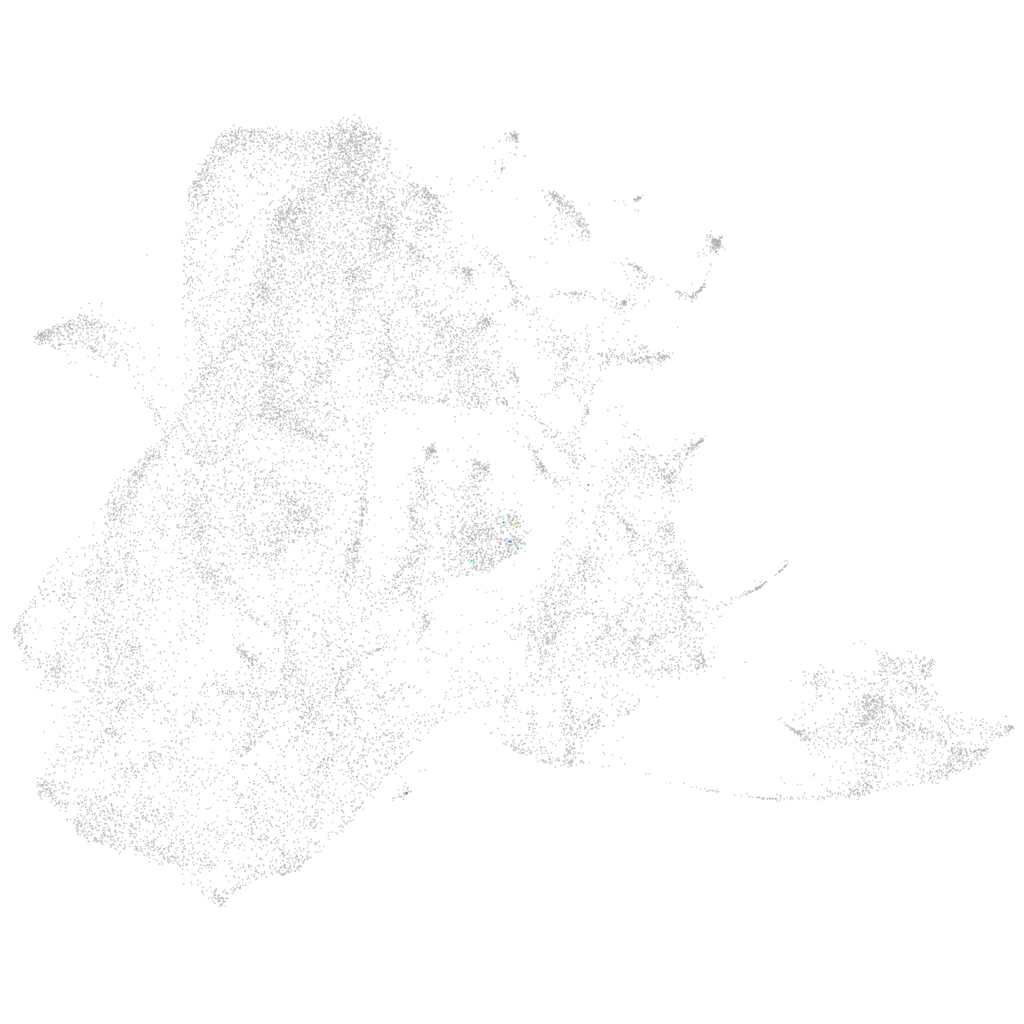
Other cell groups


all cells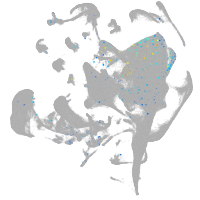 |
blastomeres |
PGCs |
endoderm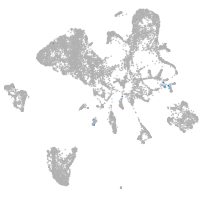 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia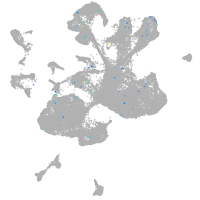 |
eye |
taste / olfactory |
otic / lateral line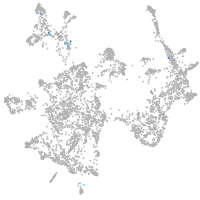 |
epidermis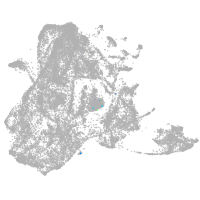 |
periderm |
fin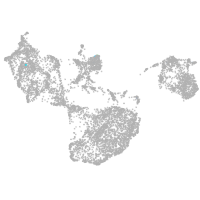 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101886653 | 0.453 | rps12 | -0.044 |
| CU019662.1 | 0.394 | rps11 | -0.043 |
| thsd7ab | 0.364 | rpl21 | -0.041 |
| CU571091.1 | 0.354 | rps15a | -0.040 |
| prkcg | 0.347 | rpl8 | -0.040 |
| kiss2 | 0.316 | rpl28 | -0.040 |
| prokr1b | 0.309 | pabpc1a | -0.039 |
| CU207241.1 | 0.279 | rps7 | -0.035 |
| si:dkey-28o19.1 | 0.274 | slc25a5 | -0.033 |
| ZNF521 (1 of many) | 0.268 | rpl26 | -0.033 |
| cpne8 | 0.266 | rps24 | -0.033 |
| abr | 0.265 | rps3 | -0.032 |
| abcc8b | 0.262 | rplp2l | -0.032 |
| CABZ01065328.1 | 0.259 | cfl1l | -0.032 |
| ephx4 | 0.239 | pfn1 | -0.030 |
| cldn2 | 0.238 | epcam | -0.029 |
| slc8a4a | 0.233 | serbp1a | -0.028 |
| XLOC-001602 | 0.231 | rps20 | -0.027 |
| chata | 0.229 | rps15 | -0.027 |
| chgb | 0.220 | rpl23 | -0.027 |
| diras2 | 0.217 | rps26 | -0.027 |
| olig3 | 0.216 | myl12.1 | -0.026 |
| si:ch73-223f5.1 | 0.216 | rpl18 | -0.026 |
| rimbp2 | 0.212 | tpt1 | -0.026 |
| zgc:152904 | 0.207 | rpl29 | -0.025 |
| igflr1 | 0.207 | ccng1 | -0.025 |
| CR356227.1 | 0.203 | rps2 | -0.025 |
| oprd1b | 0.202 | actb2 | -0.025 |
| lrrc38b | 0.201 | rpl36a | -0.025 |
| nlgn1 | 0.199 | edf1 | -0.024 |
| tnfaip2b | 0.199 | rpl11 | -0.024 |
| GPR45 | 0.195 | actb1 | -0.024 |
| LOC101885930 | 0.194 | rpl15 | -0.023 |
| nmbr | 0.192 | rpl13 | -0.023 |
| si:ch211-248l17.3 | 0.189 | myh9a | -0.023 |