DIRAS family GTPase 2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 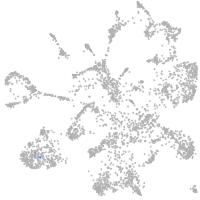 |
mesenchyme 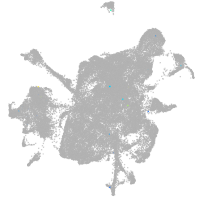 |
hematopoietic / vasculature 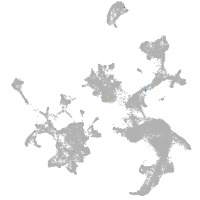 |
pronephros  |
neural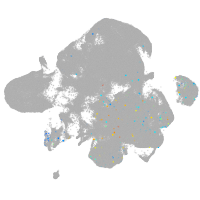 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.061 | aldob | -0.025 |
| zgc:65894 | 0.057 | id1 | -0.024 |
| elavl4 | 0.056 | banf1 | -0.023 |
| sncb | 0.056 | ccng1 | -0.022 |
| stmn1b | 0.056 | hspb1 | -0.022 |
| elavl3 | 0.055 | lig1 | -0.022 |
| mllt11 | 0.055 | mki67 | -0.022 |
| stmn2a | 0.055 | ccnd1 | -0.021 |
| rtn1a | 0.053 | chaf1a | -0.021 |
| rtn1b | 0.053 | eif4ebp3l | -0.021 |
| ywhag2 | 0.053 | fbl | -0.021 |
| ywhah | 0.053 | hdlbpa | -0.021 |
| si:dkey-276j7.1 | 0.052 | tpx2 | -0.021 |
| tuba1c | 0.052 | akap12b | -0.020 |
| vamp2 | 0.052 | ccna2 | -0.020 |
| stxbp1a | 0.052 | dkc1 | -0.020 |
| gng2 | 0.051 | fbxo5 | -0.020 |
| snap25a | 0.051 | msna | -0.020 |
| stx1b | 0.051 | nop58 | -0.020 |
| tubb5 | 0.051 | pcna | -0.020 |
| cnrip1a | 0.050 | anp32b | -0.019 |
| gap43 | 0.049 | dek | -0.019 |
| atp6v0cb | 0.048 | npm1a | -0.019 |
| fez1 | 0.048 | polr3gla | -0.019 |
| gdi1 | 0.048 | pou5f3 | -0.019 |
| id4 | 0.048 | si:ch211-152c2.3 | -0.019 |
| si:dkeyp-75h12.5 | 0.048 | stm | -0.019 |
| tmsb | 0.048 | tuba8l | -0.019 |
| calm1a | 0.048 | zgc:110425 | -0.019 |
| cdk5r1b | 0.047 | apoc1 | -0.018 |
| dpysl3 | 0.047 | apoeb | -0.018 |
| map1aa | 0.047 | asb11 | -0.018 |
| cplx2l | 0.046 | bzw1b | -0.018 |
| gnao1a | 0.046 | ccnb1 | -0.018 |
| rnasekb | 0.046 | cdca7a | -0.018 |




