si:ch73-116o1.2
ZFIN 
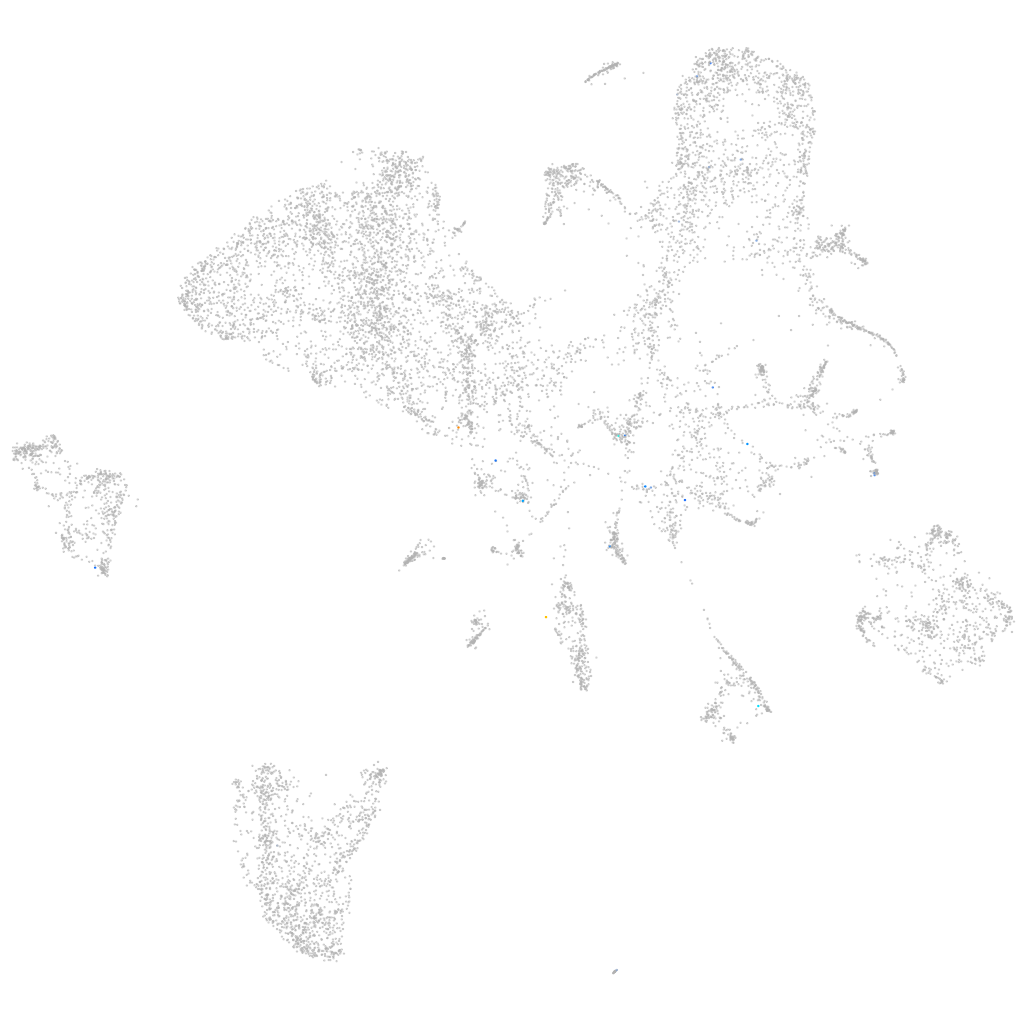
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01088025.1 | 0.554 | cox6a1 | -0.035 |
| lrrc7 | 0.511 | COX5B | -0.035 |
| LOC100334612 | 0.482 | gapdh | -0.034 |
| LOC101884484 | 0.435 | rps3 | -0.034 |
| tmbim1b | 0.370 | si:ch1073-325m22.2 | -0.034 |
| XLOC-001430 | 0.362 | suclg1 | -0.033 |
| si:ch211-62a1.3 | 0.361 | ahcy | -0.031 |
| gpr141 | 0.337 | dap | -0.031 |
| hlx1 | 0.337 | cox6c | -0.030 |
| agtr1b | 0.334 | atp5po | -0.029 |
| BX248082.1 | 0.323 | nme2b.1 | -0.029 |
| npr1a | 0.321 | gpx4a | -0.028 |
| adgra2 | 0.318 | tpt1 | -0.028 |
| CR626886.1 | 0.315 | gatm | -0.028 |
| vegfc | 0.303 | tpi1b | -0.028 |
| myct1a | 0.299 | uqcrh | -0.028 |
| XLOC-002639 | 0.299 | atp5if1b | -0.027 |
| ebf1b | 0.295 | ndufb3 | -0.027 |
| frem1a | 0.294 | nupr1b | -0.027 |
| hhat | 0.288 | rplp2 | -0.026 |
| dsela | 0.279 | aldob | -0.026 |
| fli1b | 0.278 | mat1a | -0.026 |
| drc3 | 0.269 | agxtb | -0.025 |
| XLOC-043043 | 0.267 | eno3 | -0.025 |
| rem1 | 0.253 | suclg2 | -0.025 |
| eva1bb | 0.251 | kng1 | -0.024 |
| cops6 | 0.251 | glud1b | -0.024 |
| BX901920.1 | 0.247 | tfa | -0.024 |
| flt1 | 0.247 | gamt | -0.024 |
| arhgap31 | 0.246 | atp5pd | -0.024 |
| pdgfrb | 0.231 | cox7c | -0.024 |
| entpd1 | 0.229 | rpl23 | -0.024 |
| pecam1 | 0.229 | BX908782.3 | -0.024 |
| si:ch211-162i8.4 | 0.227 | sec61b | -0.024 |
| si:ch211-246m6.5 | 0.225 | aqp12 | -0.024 |