Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 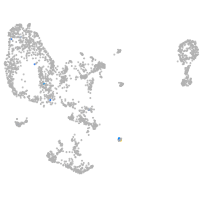 |
neural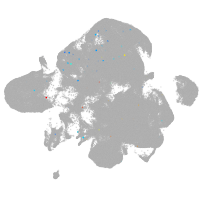 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line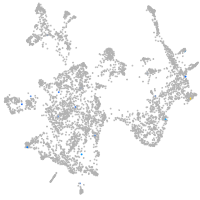 |
epidermis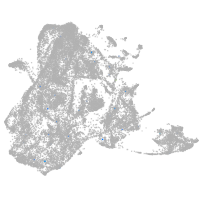 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-214p16.1 | 0.065 | pmp22a | -0.011 |
| csf1rb | 0.055 | col1a2 | -0.010 |
| rasal3 | 0.055 | col1a1a | -0.009 |
| cytip | 0.053 | col1a1b | -0.009 |
| CU499330.1 | 0.045 | CU929237.1 | -0.009 |
| lpar5a | 0.045 | fmoda | -0.009 |
| si:ch73-208g10.1 | 0.044 | col5a1 | -0.009 |
| wasa | 0.044 | mfap2 | -0.009 |
| tagapb | 0.044 | col11a1b | -0.008 |
| itgae.2 | 0.043 | col5a2a | -0.008 |
| si:dkey-185m8.2 | 0.043 | cpn1 | -0.008 |
| CU019662.1 | 0.042 | fibina | -0.008 |
| sla2 | 0.041 | mxra8b | -0.008 |
| spi2 | 0.041 | tgfbi | -0.008 |
| wasb | 0.041 | twist1a | -0.008 |
| ccr9a | 0.040 | twist1b | -0.008 |
| lcp2a | 0.040 | vwde | -0.008 |
| ccl38.6 | 0.039 | cd248b | -0.007 |
| laptm5 | 0.039 | col12a1b | -0.007 |
| si:ch211-214p16.2 | 0.039 | col2a1b | -0.007 |
| si:dkeyp-68b7.12 | 0.039 | col6a1 | -0.007 |
| grap2a | 0.038 | col6a2 | -0.007 |
| LOC100151196 | 0.037 | col6a3 | -0.007 |
| rgs13 | 0.037 | col6a4a | -0.007 |
| si:rp71-68n21.9 | 0.037 | dcn | -0.007 |
| si:dkey-192k22.2 | 0.036 | efemp2a | -0.007 |
| unc13d | 0.036 | fkbp7 | -0.007 |
| cd7al | 0.035 | hmgb3a | -0.007 |
| hmha1b | 0.035 | lpar1 | -0.007 |
| coro1a | 0.034 | meox1 | -0.007 |
| fmnl1a | 0.034 | mmp14a | -0.007 |
| ikzf1 | 0.034 | ms4a17a.11 | -0.007 |
| psmb8a | 0.034 | pdgfrl | -0.007 |
| rgs18 | 0.034 | postnb | -0.007 |
| zgc:152774 | 0.034 | reck | -0.007 |




