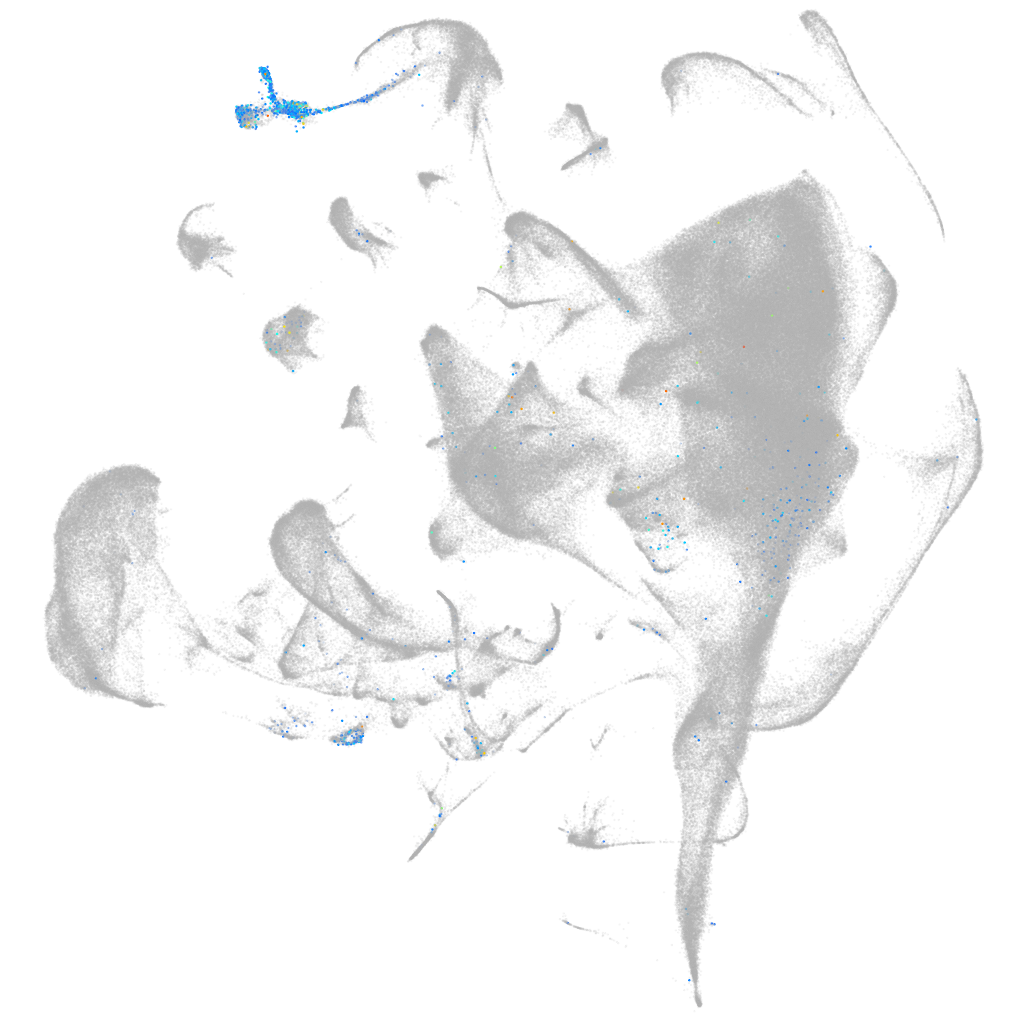
unc-13 homolog D (C. elegans)
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| laptm5 | 0.418 | mdka | -0.046 |
| coro1a | 0.396 | hmgb1b | -0.045 |
| si:dkey-185m8.2 | 0.395 | hmgb3a | -0.043 |
| itgae.2 | 0.382 | pfn2l | -0.042 |
| grap2b | 0.381 | fabp3 | -0.038 |
| lpar5a | 0.377 | jpt1b | -0.037 |
| wasa | 0.374 | sparc | -0.037 |
| wasb | 0.370 | tuba1c | -0.036 |
| plek | 0.367 | id1 | -0.035 |
| glipr1a | 0.365 | marcksb | -0.035 |
| tnfaip8l2a | 0.364 | si:ch211-137a8.4 | -0.035 |
| ptprc | 0.360 | si:ch73-46j18.5 | -0.035 |
| lcp1 | 0.357 | tspan7 | -0.035 |
| FERMT3 (1 of many) | 0.349 | hnrnpa0a | -0.034 |
| parvg | 0.344 | nova2 | -0.034 |
| nrros | 0.343 | tmeff1b | -0.034 |
| myo1f | 0.341 | tuba1a | -0.034 |
| itgb2 | 0.340 | her15.1 | -0.034 |
| rgs13 | 0.340 | si:ch211-133n4.4 | -0.033 |
| si:ch73-248e21.5 | 0.340 | pmp22a | -0.032 |
| vaspa | 0.340 | si:dkey-56m19.5 | -0.032 |
| zgc:64051 | 0.340 | cx43.4 | -0.031 |
| ccr9a | 0.332 | fabp7a | -0.031 |
| si:ch211-214p16.1 | 0.332 | pbx4 | -0.031 |
| si:rp71-68n21.9 | 0.331 | mfap2 | -0.031 |
| spi1b | 0.328 | gpm6aa | -0.030 |
| hmha1b | 0.327 | sox11a | -0.030 |
| si:dkeyp-68b7.12 | 0.323 | sox11b | -0.030 |
| CU499336.2 | 0.322 | elavl3 | -0.029 |
| si:dkey-102g19.3 | 0.317 | fam168a | -0.029 |
| si:dkey-27h10.2 | 0.317 | foxp4 | -0.029 |
| arhgap15 | 0.316 | fxyd6l | -0.029 |
| fmnl1a | 0.315 | tubb5 | -0.029 |
| gmfg | 0.315 | angptl4 | -0.028 |
| alox5ap | 0.314 | cdh2 | -0.028 |