si:ch211-66e2.5
ZFIN 
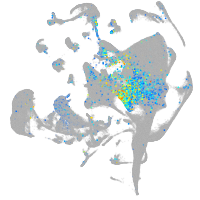
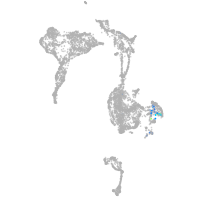
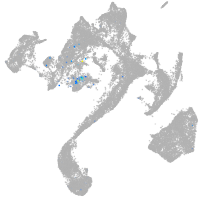
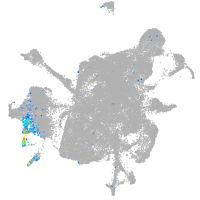
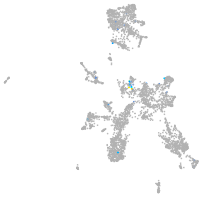
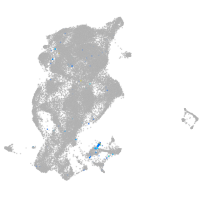
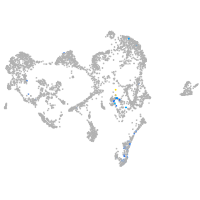
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp1a1b | 0.423 | and1 | -0.067 |
| zgc:165461 | 0.396 | CABZ01074130.1 | -0.059 |
| hepacama | 0.393 | col1a1a | -0.058 |
| slc6a11b | 0.355 | LOC100538221 | -0.051 |
| atp1b4 | 0.343 | and2 | -0.045 |
| slc1a3b | 0.338 | msx2b | -0.043 |
| ccdc170 | 0.329 | zgc:66479 | -0.040 |
| slc1a2b | 0.293 | lrrc17 | -0.039 |
| gpr37l1b | 0.283 | si:ch211-264f5.6 | -0.039 |
| fabp7a | 0.260 | kank3 | -0.038 |
| LOC101882465 | 0.259 | im:7150988 | -0.037 |
| slc3a2a | 0.250 | il17rd | -0.036 |
| adra1ab | 0.248 | kctd5a | -0.035 |
| masp1 | 0.243 | colgalt2 | -0.035 |
| LOC110437924.1 | 0.241 | krt97 | -0.034 |
| CU467822.1 | 0.236 | dlx2b | -0.034 |
| her8.2 | 0.233 | fmodb | -0.034 |
| si:ch211-251b21.1 | 0.232 | kif26ab | -0.033 |
| LOC103910690 | 0.224 | angptl7 | -0.033 |
| slc47a4 | 0.221 | NC-002333.4 | -0.033 |
| cfap157 | 0.221 | acy3.2 | -0.032 |
| cldn7a | 0.218 | dusp6 | -0.032 |
| zgc:109982 | 0.218 | capn12 | -0.031 |
| slc4a4a | 0.217 | plscr3b | -0.031 |
| hspb15 | 0.216 | etv4 | -0.031 |
| abi3a | 0.215 | sp9 | -0.031 |
| si:ch73-265d7.2 | 0.215 | bambia | -0.030 |
| robo4 | 0.215 | hoxa11b | -0.030 |
| gdf5 | 0.214 | sec61g | -0.030 |
| msi1 | 0.212 | arid3c | -0.030 |
| efhd1 | 0.211 | vwa2 | -0.029 |
| BX119909.1 | 0.210 | vcanb | -0.029 |
| glula | 0.210 | fras1 | -0.029 |
| eno1b | 0.209 | matn4 | -0.029 |
| cldn5a | 0.208 | klf3 | -0.028 |