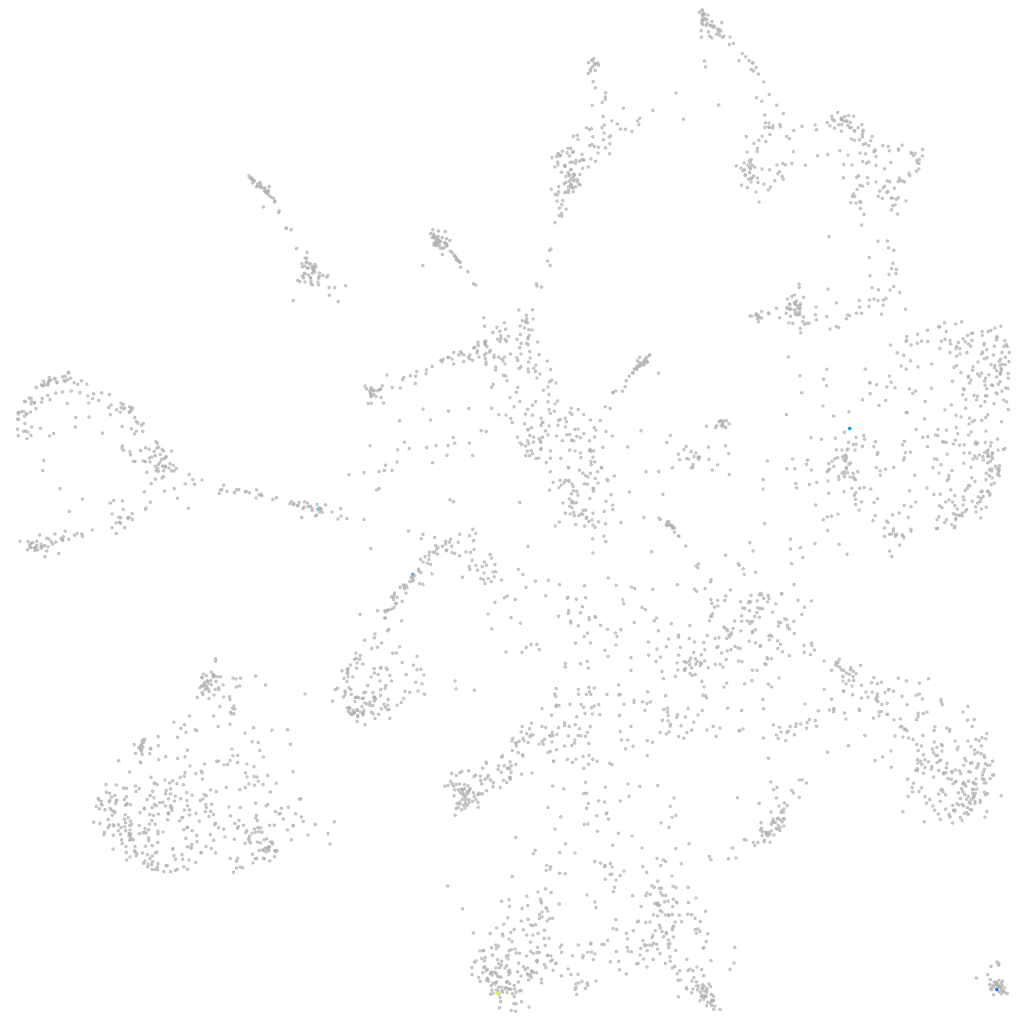
si:ch211-219a15.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| foxe3 | 0.652 | rps18 | -0.035 |
| crygmxl2 | 0.652 | hmga2 | -0.031 |
| gja8a | 0.652 | cfd | -0.030 |
| CR394572.1 | 0.562 | csnk1a1 | -0.030 |
| CR356246.1 | 0.516 | hmgb3a | -0.028 |
| bspry | 0.463 | qkia | -0.027 |
| si:dkey-246j7.1 | 0.407 | si:dkey-261h17.1 | -0.027 |
| slc7a11 | 0.385 | rps11 | -0.026 |
| LOC101885950 | 0.371 | ywhaqb | -0.026 |
| col28a1a | 0.364 | sept7a | -0.026 |
| tent5d | 0.361 | ddx61 | -0.026 |
| gja8b | 0.360 | rps27.1 | -0.025 |
| wnt7ba | 0.332 | fabp3 | -0.025 |
| fev | 0.322 | lsm4 | -0.025 |
| CU929365.1 | 0.312 | si:ch73-46j18.5 | -0.025 |
| si:ch211-133l5.7 | 0.282 | ilf3b | -0.024 |
| sparcl1 | 0.266 | map1lc3b | -0.024 |
| acsl2 | 0.265 | si:ch211-286o17.1 | -0.024 |
| si:dkey-30k22.7 | 0.265 | si:dkey-56m19.5 | -0.023 |
| LOC100007824 | 0.250 | hmgn7 | -0.023 |
| LOC103909452 | 0.249 | mtdha | -0.023 |
| cx28.9 | 0.249 | kctd12.2 | -0.023 |
| zgc:112334 | 0.220 | itga1 | -0.022 |
| mipb | 0.211 | marcksb | -0.022 |
| BFSP1 | 0.193 | pdia3 | -0.022 |
| nfil3-6 | 0.188 | gsta.1 | -0.022 |
| lrrc9 | 0.187 | mdkb | -0.022 |
| ccr9a | 0.186 | lama4 | -0.022 |
| tnxba | 0.178 | ctbp1 | -0.021 |
| crybb1 | 0.176 | rhag | -0.021 |
| tspan33b | 0.171 | wbp2nl | -0.021 |
| znf1168 | 0.167 | zgc:158463 | -0.021 |
| pde6c | 0.165 | med11 | -0.021 |
| crybb1l2 | 0.165 | chd4a | -0.021 |
| csf3b | 0.162 | smarca4a | -0.021 |