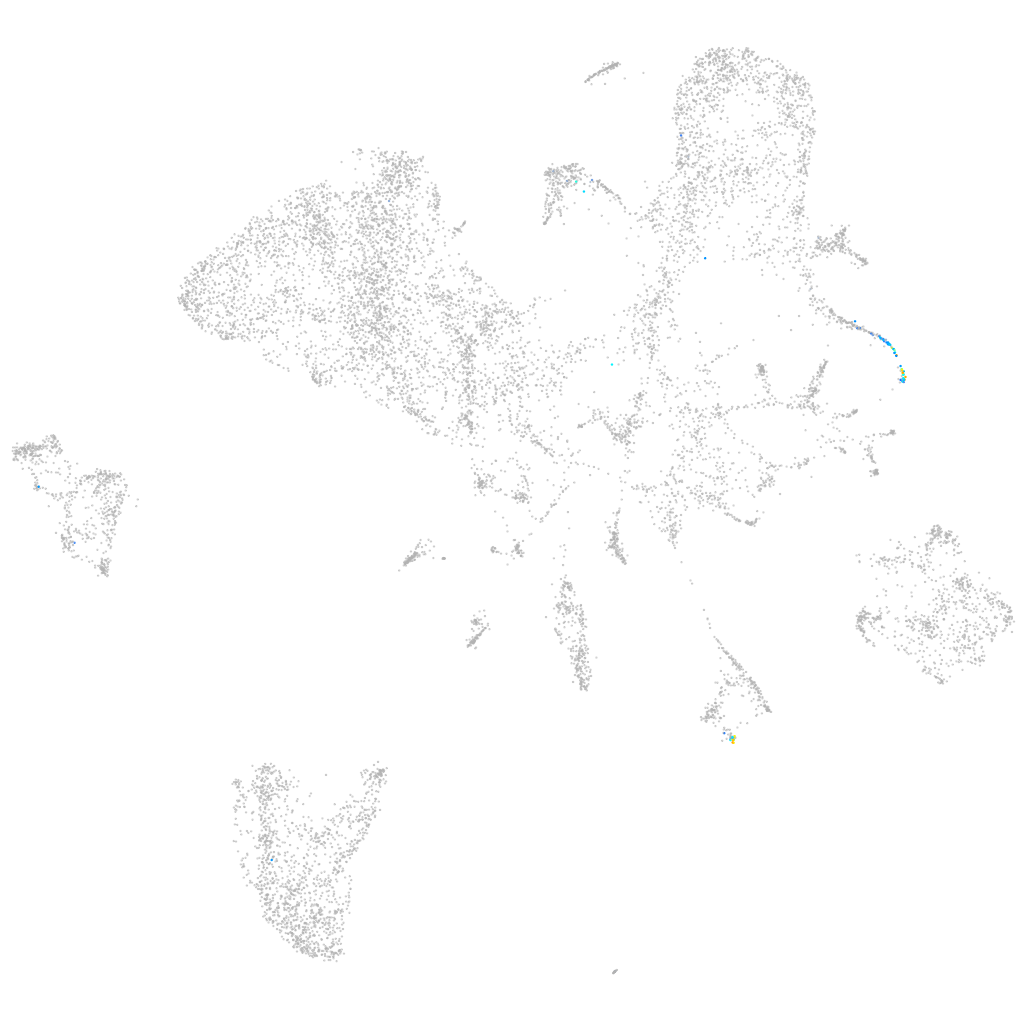
si:ch211-153b23.3
ZFIN 
Other cell groups


all cells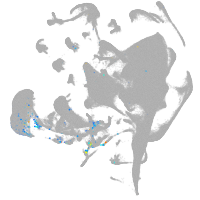 |
blastomeres |
PGCs |
endoderm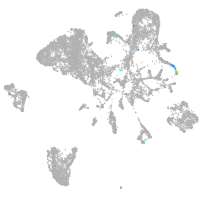 |
axial mesoderm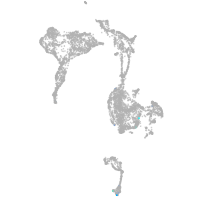 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 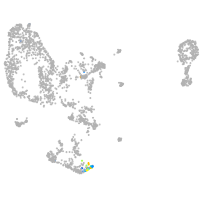 |
neural |
spinal cord / glia |
eye |
taste / olfactory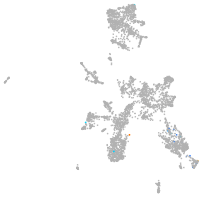 |
otic / lateral line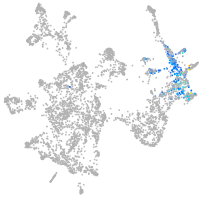 |
epidermis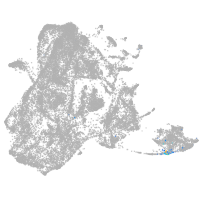 |
periderm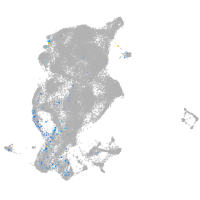 |
fin |
ionocytes / mucous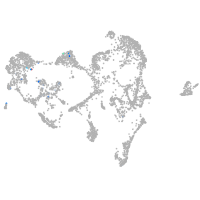 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| irg1l | 0.848 | ahcy | -0.080 |
| si:ch211-153b23.4 | 0.679 | gapdh | -0.073 |
| zgc:174917 | 0.611 | gamt | -0.071 |
| muc5e | 0.538 | gatm | -0.071 |
| mslna | 0.434 | bhmt | -0.067 |
| sp8a | 0.421 | fbp1b | -0.065 |
| evx1 | 0.379 | gstt1a | -0.063 |
| tmem176l.4 | 0.366 | apoc2 | -0.062 |
| msx2b | 0.365 | apoa4b.1 | -0.062 |
| ccl20a.3 | 0.352 | mat1a | -0.062 |
| LOC795897 | 0.335 | apoa1b | -0.062 |
| tnfrsf9a | 0.330 | afp4 | -0.060 |
| si:ch211-153b23.5 | 0.329 | hspe1 | -0.059 |
| LOC101882926 | 0.321 | aqp12 | -0.058 |
| etf1a | 0.308 | scp2a | -0.058 |
| ca4c | 0.304 | si:dkey-16p21.8 | -0.057 |
| trhr2 | 0.303 | mdh1aa | -0.057 |
| dlx2b | 0.294 | grhprb | -0.056 |
| SBSPON | 0.294 | dap | -0.056 |
| tmprss2 | 0.294 | gpx4a | -0.056 |
| ttc22 | 0.291 | sult2st2 | -0.055 |
| gphb5 | 0.289 | shmt1 | -0.055 |
| noxo1b | 0.281 | cdo1 | -0.055 |
| hoxd13a | 0.279 | apoc1 | -0.055 |
| saa | 0.278 | gpt2l | -0.054 |
| si:dkey-92i15.4 | 0.278 | dhrs9 | -0.054 |
| cxcl8a | 0.275 | glud1b | -0.054 |
| prdm1a | 0.275 | gstr | -0.054 |
| tspan35 | 0.273 | apobb.1 | -0.052 |
| rnf183 | 0.254 | gpd1b | -0.052 |
| klf2a | 0.248 | agxtb | -0.052 |
| soul2 | 0.243 | abat | -0.052 |
| CR855311.1 | 0.243 | pnp4b | -0.052 |
| CR407590.2 | 0.242 | rdh1 | -0.052 |
| CABZ01067151.1 | 0.238 | gcshb | -0.052 |