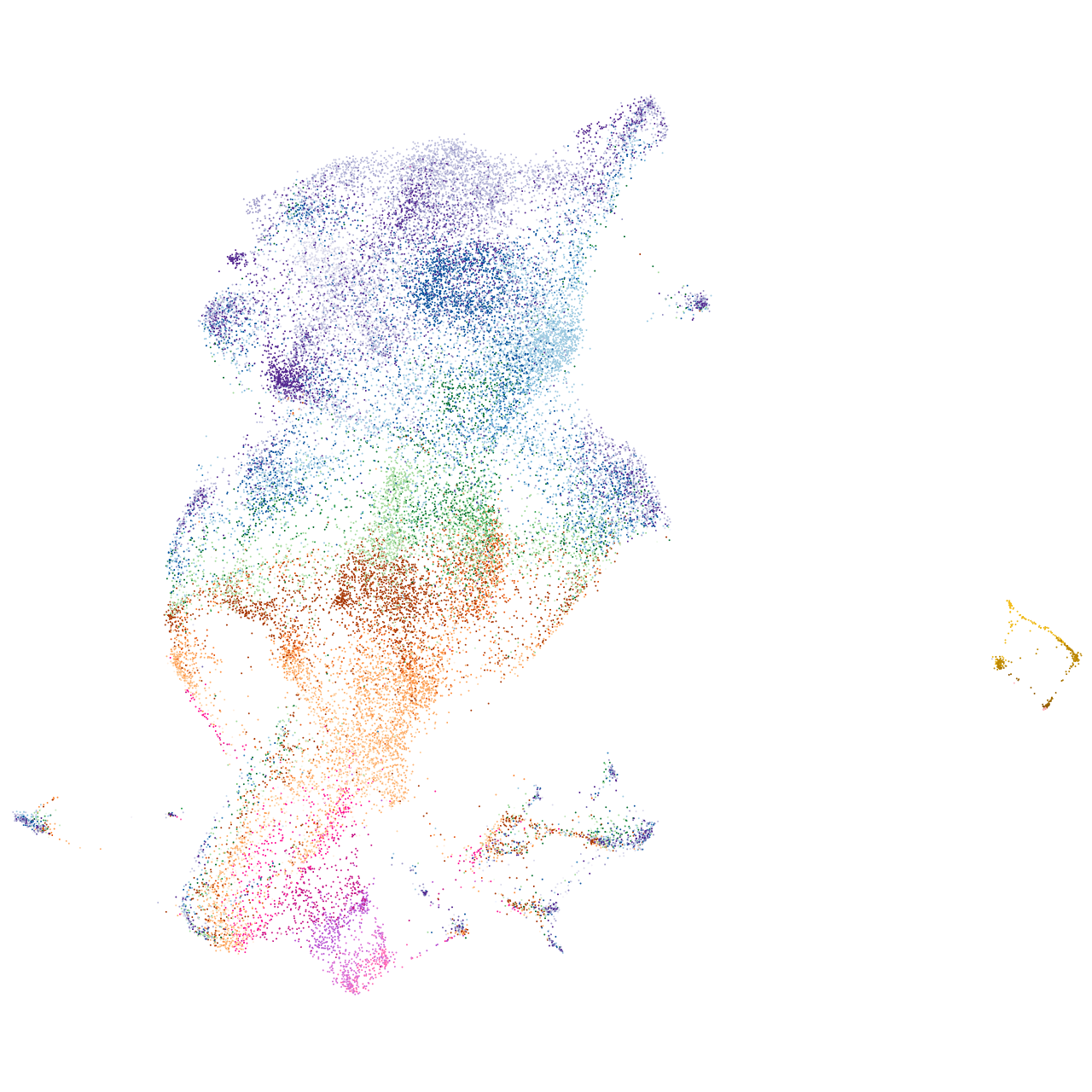si:ch211-149l1.2
ZFIN 
Other cell groups


all cells |
blastomeres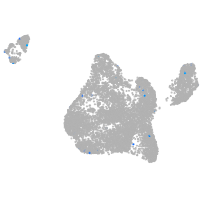 |
PGCs |
endoderm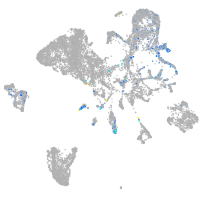 |
axial mesoderm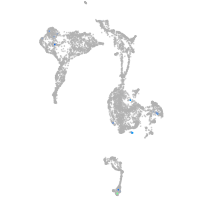 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia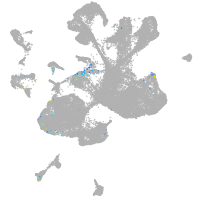 |
eye |
taste / olfactory |
otic / lateral line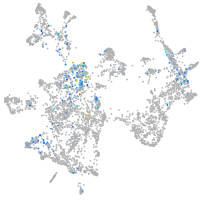 |
epidermis |
periderm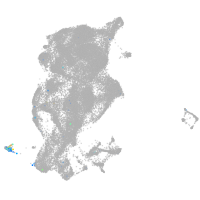 |
fin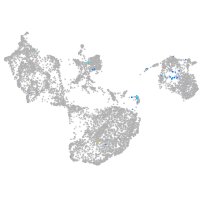 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR759836.1 | 0.408 | gsta.1 | -0.030 |
| grp | 0.402 | cyt1 | -0.030 |
| CABZ01073083.1 | 0.343 | si:ch211-105c13.3 | -0.028 |
| gfral | 0.322 | s100v1 | -0.027 |
| si:ch211-256e16.6 | 0.312 | pycard | -0.027 |
| pvalb8 | 0.293 | rpl26 | -0.027 |
| cldnh | 0.291 | aldob | -0.026 |
| vill | 0.276 | rps3a | -0.026 |
| prr15la | 0.265 | mapre1a | -0.025 |
| cldn8 | 0.250 | krt4 | -0.025 |
| fndc1 | 0.240 | krt17 | -0.024 |
| hepacam2 | 0.231 | fxyd6l | -0.024 |
| zgc:101744 | 0.211 | ywhabl | -0.024 |
| CABZ01095001.1 | 0.211 | eef1a1l1 | -0.024 |
| wdr95 | 0.206 | capgb | -0.024 |
| BX247868.1 | 0.191 | cfl1l | -0.024 |
| tyrobp | 0.188 | rpl11 | -0.023 |
| spef2 | 0.186 | rpl18 | -0.023 |
| stom | 0.185 | actb2 | -0.023 |
| chga | 0.178 | eif3ba | -0.022 |
| otofa | 0.175 | krt5 | -0.022 |
| cers3b | 0.174 | rps4x | -0.022 |
| CR812468.1 | 0.173 | pof1b | -0.022 |
| syngr2b | 0.168 | smim13 | -0.022 |
| tmprss2 | 0.164 | mgst1.2 | -0.022 |
| pcbp3 | 0.162 | si:ch73-52f15.5 | -0.022 |
| BX649499.1 | 0.162 | cct5 | -0.021 |
| eya1 | 0.161 | vdac2 | -0.021 |
| itpr3 | 0.160 | si:ch211-210c8.7 | -0.021 |
| si:ch211-207j7.2 | 0.158 | si:ch211-207n23.2 | -0.021 |
| mmp17b | 0.154 | si:dkey-222n6.2 | -0.021 |
| dmrta2 | 0.153 | si:ch1073-325m22.2 | -0.021 |
| si:ch211-247i17.1 | 0.149 | slc25a5 | -0.021 |
| penkb | 0.148 | reep5 | -0.021 |
| foxa1 | 0.148 | selenow2b | -0.021 |