si:ch211-11c3.9
ZFIN 
Other cell groups


all cells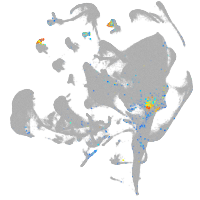 |
blastomeres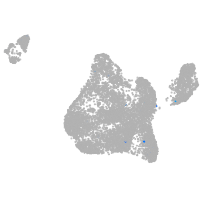 |
PGCs |
endoderm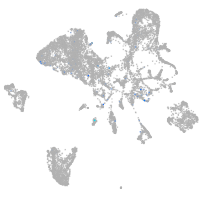 |
axial mesoderm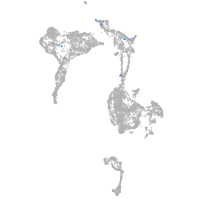 |
muscle 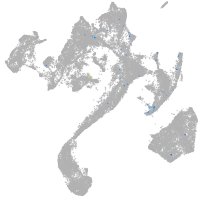 |
mural cells / non-skeletal muscle  |
mesenchyme 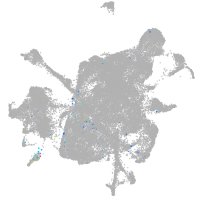 |
hematopoietic / vasculature 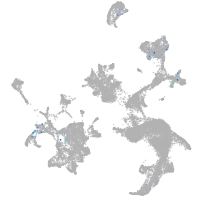 |
pronephros 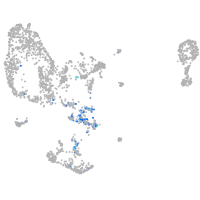 |
neural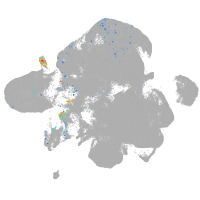 |
spinal cord / glia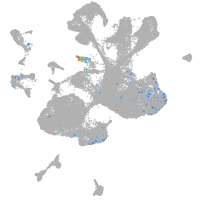 |
eye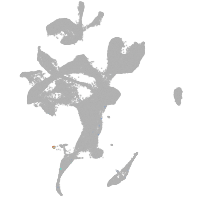 |
taste / olfactory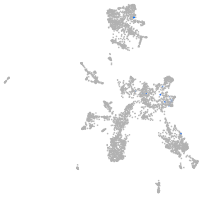 |
otic / lateral line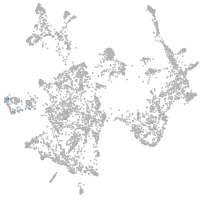 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-11c3.12 | 0.510 | gpm6aa | -0.091 |
| ednrab | 0.462 | ckbb | -0.083 |
| tfec | 0.377 | nova2 | -0.078 |
| snai1b | 0.344 | rtn1a | -0.073 |
| crestin | 0.325 | fabp7a | -0.068 |
| BX248318.1 | 0.249 | COX3 | -0.068 |
| si:ch211-11c3.11 | 0.248 | pvalb1 | -0.068 |
| sfrp2 | 0.208 | tmsb | -0.067 |
| inka1a | 0.196 | CU467822.1 | -0.067 |
| si:dkey-250k10.4 | 0.188 | elavl3 | -0.066 |
| nradd | 0.181 | CR383676.1 | -0.065 |
| XLOC-001603 | 0.179 | tuba1c | -0.064 |
| XLOC-042222 | 0.177 | pvalb2 | -0.063 |
| LOC110438542 | 0.173 | rnasekb | -0.062 |
| BX927258.1 | 0.170 | gpm6ab | -0.062 |
| crlf1a | 0.163 | stmn1b | -0.059 |
| LOC110439533 | 0.162 | hbbe1.3 | -0.057 |
| msx1b | 0.161 | gapdhs | -0.056 |
| slc15a2 | 0.153 | actc1b | -0.056 |
| hspb1 | 0.149 | myt1b | -0.055 |
| npm1a | 0.146 | slc1a2b | -0.055 |
| dlc1 | 0.146 | CU634008.1 | -0.054 |
| kcnk5b | 0.143 | zgc:165461 | -0.054 |
| pdgfra | 0.143 | si:dkeyp-75h12.5 | -0.053 |
| itga9 | 0.141 | cadm3 | -0.052 |
| XLOC-039121 | 0.139 | vim | -0.051 |
| XLOC-032526 | 0.137 | hbae3 | -0.051 |
| ets1 | 0.135 | mt-co2 | -0.050 |
| erbb3b | 0.133 | gpm6bb | -0.050 |
| mov10b.1 | 0.133 | atp1a1b | -0.049 |
| loxl3a | 0.131 | dpysl2b | -0.049 |
| msx2b | 0.128 | elavl4 | -0.049 |
| dkc1 | 0.126 | gng3 | -0.048 |
| XLOC-001964 | 0.125 | fez1 | -0.047 |
| nhp2 | 0.124 | calm1a | -0.047 |




