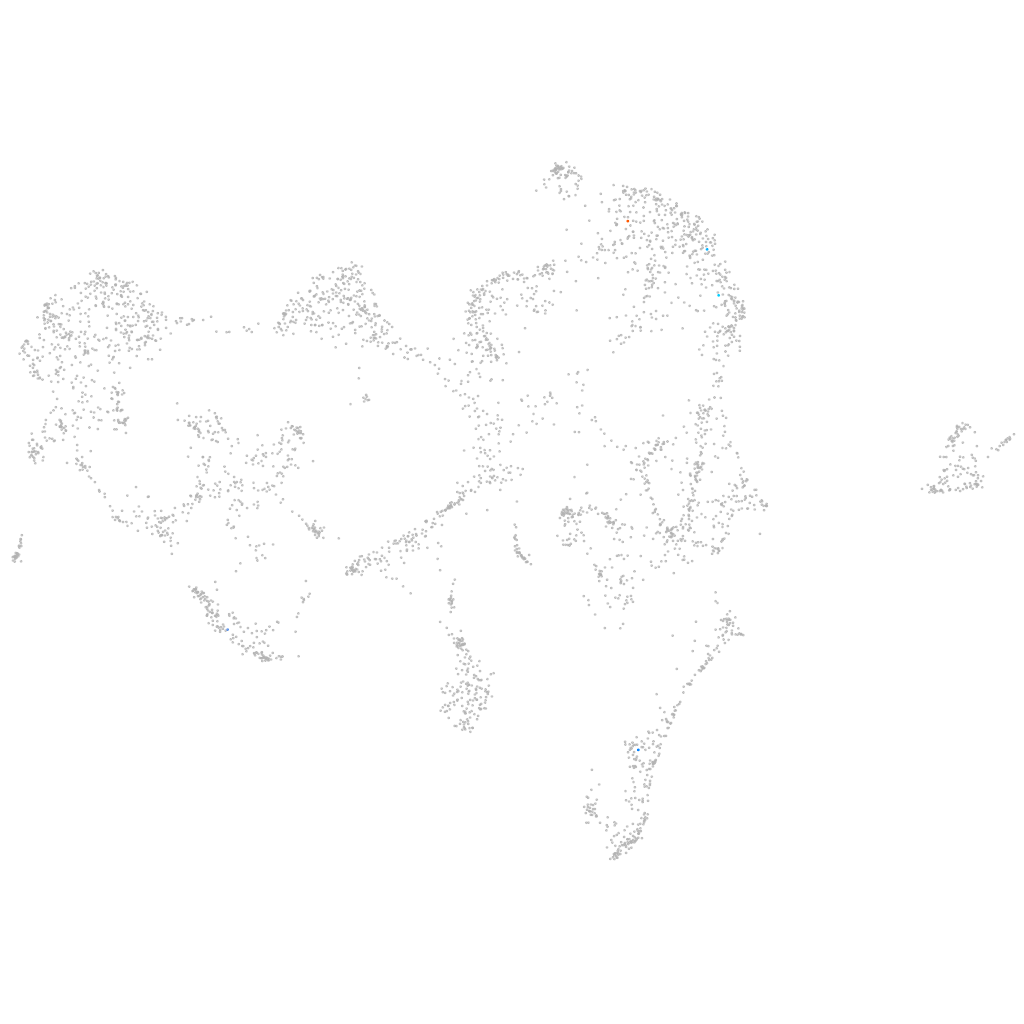
si:ch1073-513e17.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm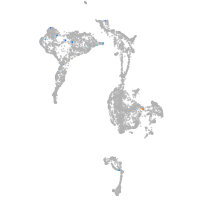 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 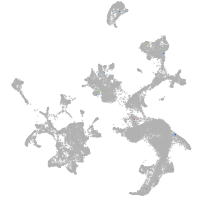 |
pronephros 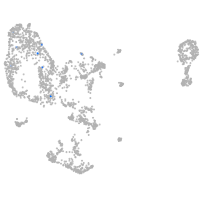 |
neural |
spinal cord / glia |
eye |
taste / olfactory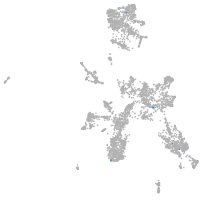 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101887126 | 0.757 | rps24 | -0.096 |
| CABZ01064248.1 | 0.675 | rpl28 | -0.093 |
| LOC103910933 | 0.594 | rpl10 | -0.080 |
| ganc | 0.505 | rpl31 | -0.075 |
| mib2 | 0.497 | rps3 | -0.056 |
| mosmob | 0.446 | rpl38 | -0.053 |
| BX322574.1 | 0.327 | rps17 | -0.050 |
| gtf2a1l | 0.316 | vdac2 | -0.050 |
| snx10b | 0.311 | rps27.2 | -0.050 |
| gas2b | 0.287 | atp5f1b | -0.048 |
| twsg1a | 0.285 | cox4i1 | -0.047 |
| srpk3 | 0.283 | atp5mc3b | -0.046 |
| esyt1a | 0.278 | cox5aa | -0.046 |
| gngt2a | 0.278 | cox6b2 | -0.045 |
| si:ch211-85n16.4 | 0.266 | eef1b2 | -0.045 |
| l1cama | 0.265 | eef1g | -0.044 |
| trim62 | 0.258 | mt-nd1 | -0.044 |
| XLOC-014039 | 0.256 | atp5l | -0.044 |
| trafd1 | 0.248 | slc25a3b | -0.043 |
| p3h4 | 0.244 | rps6 | -0.043 |
| BX088688.1 | 0.231 | ctsla | -0.043 |
| mecp2 | 0.230 | rsl24d1 | -0.042 |
| epd | 0.223 | tma7 | -0.042 |
| LOC572443 | 0.220 | atp5f1e | -0.041 |
| zgc:66455 | 0.219 | tomm20b | -0.041 |
| acot14 | 0.216 | uqcrfs1 | -0.039 |
| LOC101882575 | 0.214 | ier2a | -0.038 |
| unm-hu7910 | 0.213 | mcl1a | -0.038 |
| rbm18 | 0.209 | atp5mc1 | -0.038 |
| coq8b | 0.209 | eif1b | -0.038 |
| ttll10 | 0.208 | atp5f1c | -0.038 |
| rapgef3 | 0.205 | ran | -0.037 |
| clec16a | 0.204 | rps10 | -0.037 |
| sycn.2 | 0.204 | ndufb6 | -0.037 |
| pi4k2a | 0.202 | atp5pb | -0.037 |