Rap guanine nucleotide exchange factor (GEF) 3
ZFIN 
Other cell groups


all cells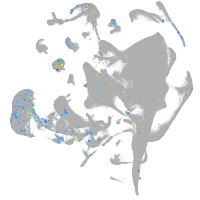 |
blastomeres |
PGCs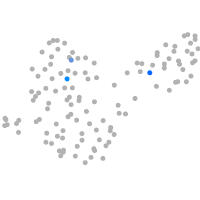 |
endoderm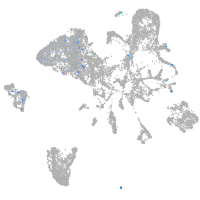 |
axial mesoderm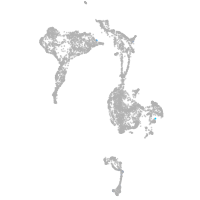 |
muscle 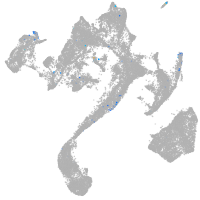 |
mural cells / non-skeletal muscle  |
mesenchyme 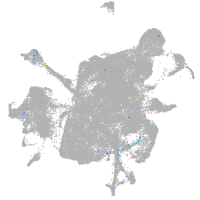 |
hematopoietic / vasculature 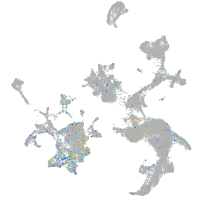 |
pronephros 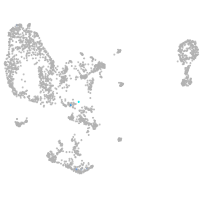 |
neural |
spinal cord / glia |
eye |
taste / olfactory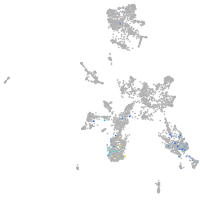 |
otic / lateral line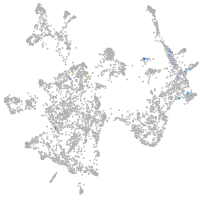 |
epidermis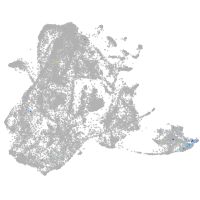 |
periderm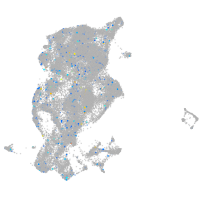 |
fin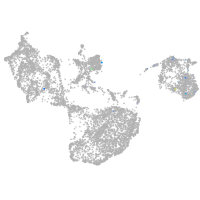 |
ionocytes / mucous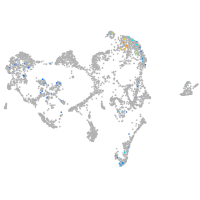 |
pigment cells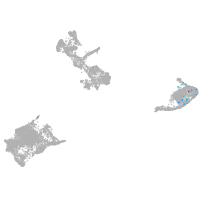 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pecam1 | 0.114 | hmgb1b | -0.055 |
| si:dkeyp-97a10.2 | 0.111 | marcksb | -0.051 |
| si:ch211-145b13.6 | 0.110 | hmgb3a | -0.050 |
| clec14a | 0.109 | si:ch211-288g17.3 | -0.049 |
| cdh5 | 0.108 | tuba1c | -0.048 |
| tie1 | 0.105 | si:ch211-137a8.4 | -0.043 |
| kdr | 0.104 | gpm6aa | -0.042 |
| she | 0.102 | mdkb | -0.042 |
| myct1a | 0.101 | zc4h2 | -0.041 |
| plvapb | 0.101 | h2afva | -0.040 |
| notchl | 0.098 | hdac1 | -0.040 |
| kdrl | 0.096 | nucks1a | -0.040 |
| flt1 | 0.094 | ckbb | -0.039 |
| rasip1 | 0.094 | smarce1 | -0.039 |
| ecscr | 0.093 | tuba1a | -0.039 |
| fgd5a | 0.086 | cbx3a | -0.038 |
| si:ch73-334d15.2 | 0.086 | fabp3 | -0.038 |
| adgrl4 | 0.085 | setb | -0.038 |
| myct1b | 0.085 | si:dkey-276j7.1 | -0.038 |
| sox7 | 0.085 | tmeff1b | -0.038 |
| si:ch211-248e11.2 | 0.084 | elavl3 | -0.037 |
| etv2 | 0.081 | hnrnpa0a | -0.037 |
| ptprb | 0.081 | hp1bp3 | -0.037 |
| slc9a3r1b | 0.081 | nova2 | -0.037 |
| plk2b | 0.080 | vdac1 | -0.037 |
| tek | 0.080 | chd4a | -0.036 |
| yrk | 0.080 | nono | -0.036 |
| plvapa | 0.078 | rtn1a | -0.036 |
| scarf1 | 0.078 | seta | -0.036 |
| ccm2l | 0.077 | cx43.4 | -0.035 |
| erg | 0.077 | ewsr1a | -0.035 |
| srgn | 0.076 | si:dkey-56m19.5 | -0.035 |
| cav1 | 0.075 | stmn1b | -0.035 |
| egfl7 | 0.075 | tubb5 | -0.035 |
| LOC101882826 | 0.075 | ddx39ab | -0.034 |




