zgc:66455
ZFIN 
Other cell groups


all cells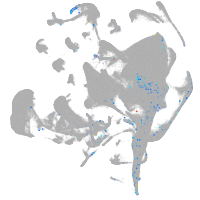 |
blastomeres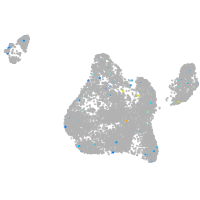 |
PGCs |
endoderm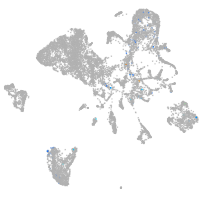 |
axial mesoderm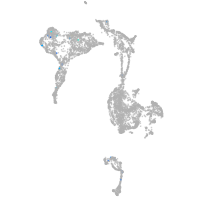 |
muscle 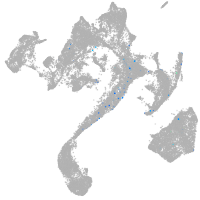 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 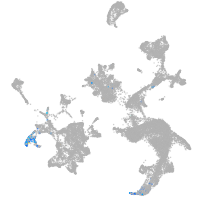 |
pronephros 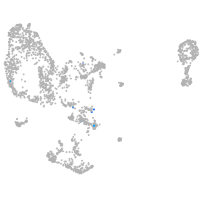 |
neural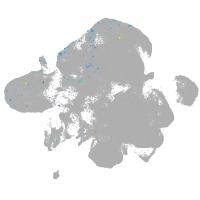 |
spinal cord / glia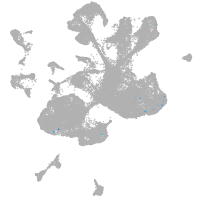 |
eye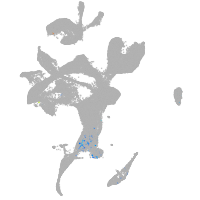 |
taste / olfactory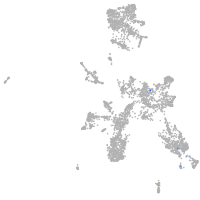 |
otic / lateral line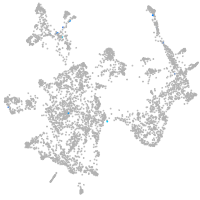 |
epidermis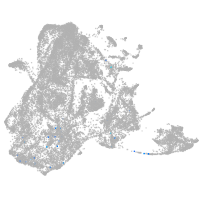 |
periderm |
fin |
ionocytes / mucous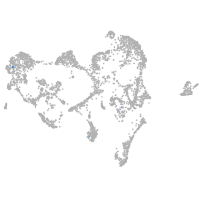 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gfi1aa | 0.089 | ccni | -0.028 |
| cldng | 0.078 | marcksl1a | -0.028 |
| npm1a | 0.078 | pvalb1 | -0.027 |
| snu13b | 0.078 | gpm6aa | -0.025 |
| si:dkey-261j4.4 | 0.075 | pvalb2 | -0.025 |
| nhp2 | 0.074 | elavl3 | -0.024 |
| dkc1 | 0.073 | krt4 | -0.023 |
| nop58 | 0.073 | sparc | -0.023 |
| ssb | 0.072 | ckbb | -0.022 |
| fbl | 0.069 | col1a2 | -0.022 |
| pprc1 | 0.069 | cyt1 | -0.022 |
| cad | 0.068 | gpm6ab | -0.022 |
| rpf2 | 0.068 | stmn1b | -0.022 |
| c1qbp | 0.067 | actc1b | -0.021 |
| mrto4 | 0.067 | krtt1c19e | -0.021 |
| nop10 | 0.067 | tmsb | -0.021 |
| pa2g4a | 0.067 | tuba1c | -0.021 |
| rpl7l1 | 0.067 | dap1b | -0.021 |
| nifk | 0.066 | anxa1a | -0.020 |
| nip7 | 0.065 | col1a1a | -0.020 |
| nop56 | 0.065 | col1a1b | -0.020 |
| prps1a | 0.065 | CU467822.1 | -0.020 |
| rcl1 | 0.065 | cyt1l | -0.020 |
| si:ch73-299h12.2 | 0.065 | hmgb3a | -0.020 |
| bxdc2 | 0.064 | pmp22a | -0.020 |
| hmbsa | 0.064 | rtn1a | -0.020 |
| morc3b | 0.064 | CU634008.1 | -0.020 |
| nop2 | 0.064 | ccng2 | -0.019 |
| rrp1 | 0.064 | icn | -0.019 |
| tomm20a | 0.064 | icn2 | -0.019 |
| tsr2 | 0.063 | ip6k2a | -0.019 |
| twistnb | 0.063 | myt1a | -0.019 |
| umps | 0.063 | myt1b | -0.019 |
| wdr43 | 0.063 | ndrg2 | -0.019 |
| znfl2a | 0.063 | nova2 | -0.019 |




