serine incorporator 2
ZFIN 
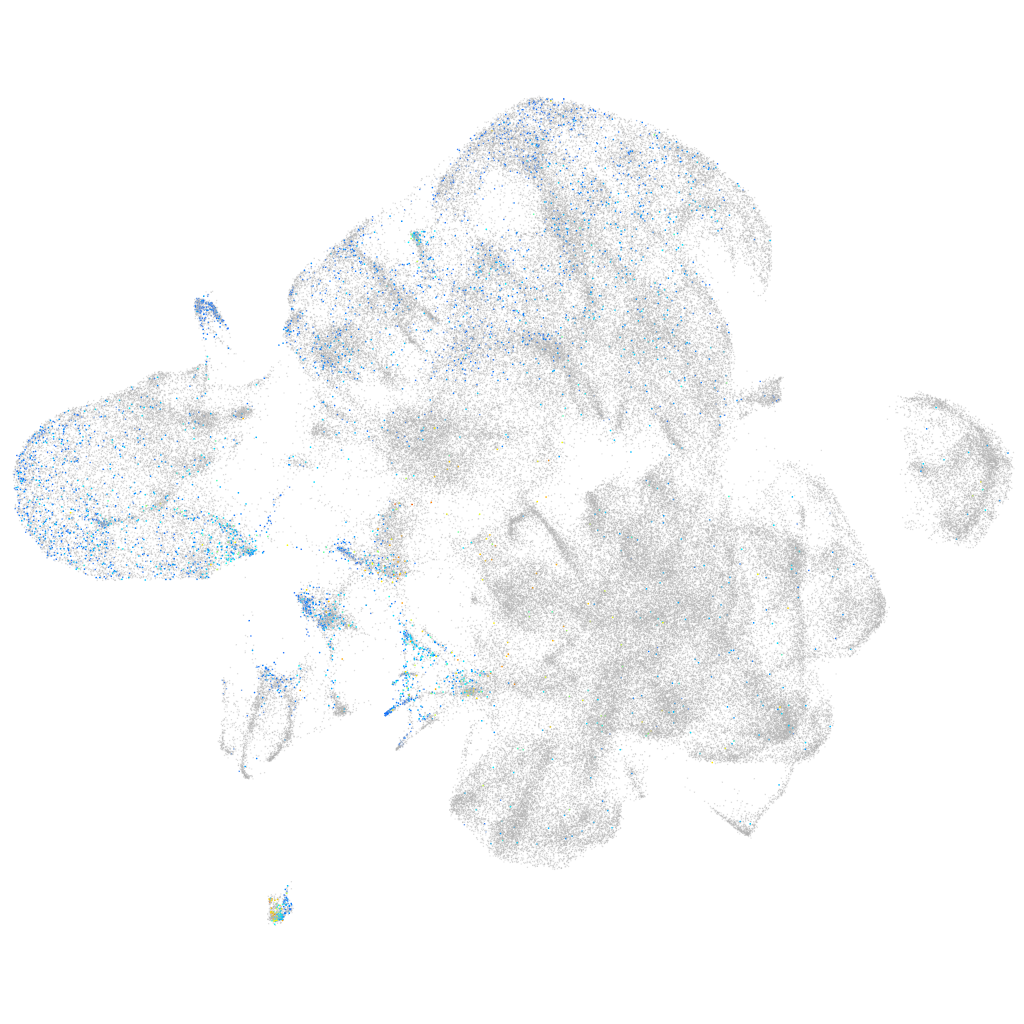
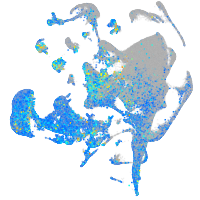
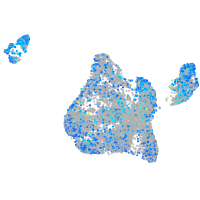
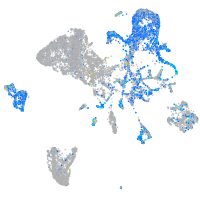
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| epcam | 0.148 | tuba1c | -0.122 |
| krt8 | 0.142 | elavl3 | -0.113 |
| cd9b | 0.116 | rtn1a | -0.109 |
| f11r.1 | 0.115 | nova2 | -0.107 |
| cldnh | 0.113 | ptmaa | -0.105 |
| grp | 0.113 | stmn1b | -0.104 |
| hopx | 0.112 | gpm6aa | -0.103 |
| spint2 | 0.111 | gng3 | -0.089 |
| ucp2 | 0.111 | tuba1a | -0.087 |
| CR759836.1 | 0.110 | tmsb | -0.086 |
| dnase1l4.1 | 0.109 | ckbb | -0.086 |
| zgc:101744 | 0.109 | tubb5 | -0.082 |
| degs2 | 0.108 | hmgb3a | -0.082 |
| apoeb | 0.107 | gpm6ab | -0.081 |
| cdh1 | 0.106 | sncb | -0.079 |
| zgc:92380 | 0.106 | rtn1b | -0.079 |
| sult2st1 | 0.105 | si:dkey-276j7.1 | -0.078 |
| anxa4 | 0.105 | elavl4 | -0.077 |
| cldn7b | 0.105 | zc4h2 | -0.077 |
| pmp22a | 0.104 | fez1 | -0.076 |
| pvalb8 | 0.104 | atp6v0cb | -0.074 |
| itpr3 | 0.104 | gng2 | -0.074 |
| gfral | 0.103 | ywhah | -0.074 |
| cldnb | 0.102 | celf2 | -0.073 |
| apoc1 | 0.102 | tmsb4x | -0.072 |
| si:ch1073-443f11.2 | 0.101 | ywhag2 | -0.072 |
| prr15la | 0.101 | myt1b | -0.072 |
| akap12b | 0.101 | stmn2a | -0.071 |
| vill | 0.100 | rnasekb | -0.070 |
| pou2f3 | 0.100 | stx1b | -0.069 |
| si:ch211-202h22.8 | 0.099 | si:dkeyp-75h12.5 | -0.069 |
| bambia | 0.099 | gnao1a | -0.069 |
| cnn2 | 0.099 | stxbp1a | -0.068 |
| krt97 | 0.099 | mllt11 | -0.068 |
| bik | 0.099 | dpysl2b | -0.068 |