selenoprotein P2
ZFIN 
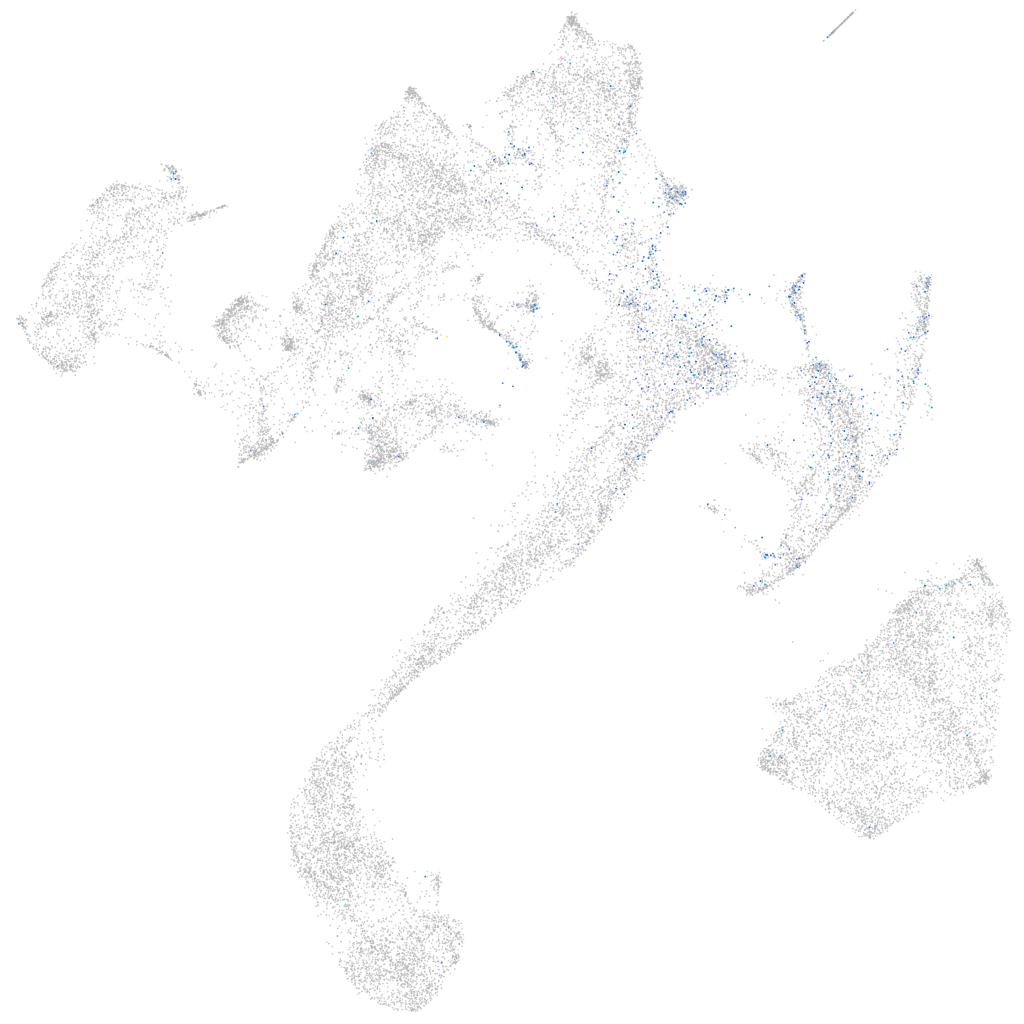
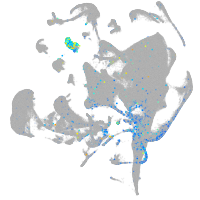
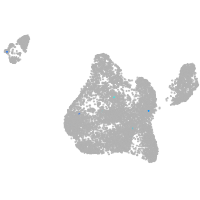
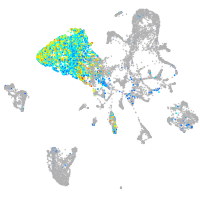
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fn1b | 0.123 | ckma | -0.067 |
| fbln1 | 0.118 | atp2a1 | -0.065 |
| kazald2 | 0.112 | ckmb | -0.064 |
| meox1 | 0.105 | gapdh | -0.064 |
| ripply1 | 0.100 | actc1b | -0.063 |
| sall4 | 0.098 | pabpc4 | -0.061 |
| XLOC-001964 | 0.096 | mylpfa | -0.061 |
| hnrnpa0l | 0.094 | tmem38a | -0.060 |
| pcdh8 | 0.092 | aldoab | -0.059 |
| lin28a | 0.092 | si:dkey-16p21.8 | -0.058 |
| draxin | 0.091 | bhmt | -0.058 |
| arid3c | 0.091 | ak1 | -0.057 |
| rasgef1ba | 0.090 | tnnt3a | -0.057 |
| LO017843.1 | 0.090 | ank1a | -0.057 |
| tuba8l2 | 0.088 | myl1 | -0.057 |
| tspan7 | 0.088 | srl | -0.056 |
| selenof | 0.087 | rplp2 | -0.056 |
| nid2a | 0.085 | mylz3 | -0.056 |
| si:ch1073-429i10.3.1 | 0.084 | pvalb1 | -0.055 |
| XLOC-042222 | 0.084 | pvalb2 | -0.055 |
| btg2 | 0.083 | cox17 | -0.055 |
| id3 | 0.082 | si:ch73-367p23.2 | -0.055 |
| ubc | 0.082 | tnnc2 | -0.054 |
| COX7A2 (1 of many) | 0.082 | pgam2 | -0.054 |
| ubxn8 | 0.081 | ldb3b | -0.054 |
| CU463284.1 | 0.081 | acta1b | -0.053 |
| zgc:56493 | 0.081 | actn3a | -0.052 |
| sipa1l2 | 0.080 | neb | -0.052 |
| ssr3 | 0.080 | tnni2a.4 | -0.052 |
| hsp90ab1 | 0.079 | myom1a | -0.052 |
| h3f3a | 0.079 | fabp3 | -0.052 |
| XLOC-042229 | 0.079 | cav3 | -0.052 |
| si:dkey-261h17.1 | 0.079 | eno1a | -0.052 |
| CR759845.4 | 0.078 | gamt | -0.052 |
| nfyba | 0.078 | CABZ01078594.1 | -0.051 |