RNA pseudouridine synthase D4
ZFIN 
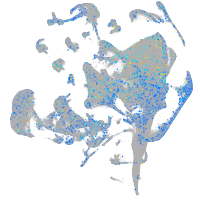
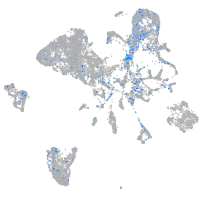
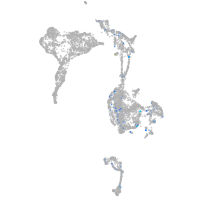
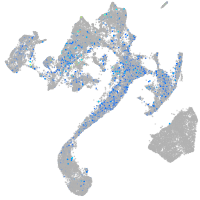
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:56493 | 0.090 | marcksl1b | -0.057 |
| dut | 0.082 | rtn1a | -0.056 |
| pa2g4a | 0.079 | gpm6aa | -0.053 |
| ftsj3 | 0.078 | elavl3 | -0.053 |
| eif6 | 0.078 | NC-002333.4 | -0.049 |
| paics | 0.078 | stmn1b | -0.047 |
| nutf2l | 0.077 | gpm6ab | -0.042 |
| selenoh | 0.077 | myt1b | -0.041 |
| snu13b | 0.077 | hmgb3a | -0.040 |
| shmt1 | 0.076 | si:dkey-56m19.5 | -0.038 |
| mcm7 | 0.076 | hmgb1a | -0.037 |
| mcm2 | 0.075 | stm | -0.037 |
| c1qbp | 0.074 | ckbb | -0.037 |
| mcm5 | 0.074 | si:dkeyp-75h12.5 | -0.037 |
| pcna | 0.074 | sncb | -0.037 |
| bysl | 0.074 | atp6v0cb | -0.037 |
| tma16 | 0.073 | CR383676.1 | -0.036 |
| pa2g4b | 0.072 | nova2 | -0.036 |
| adi1 | 0.072 | rnasekb | -0.036 |
| si:dkey-102m7.3 | 0.071 | si:ch211-152c2.3 | -0.036 |
| nop10 | 0.071 | vamp2 | -0.035 |
| mcm4 | 0.071 | tubb5 | -0.035 |
| sirt7 | 0.070 | apoc1 | -0.034 |
| rpa2 | 0.069 | myt1a | -0.034 |
| zgc:114188 | 0.069 | gng3 | -0.034 |
| rps10 | 0.068 | tp53inp2 | -0.034 |
| gart | 0.068 | pou5f3 | -0.033 |
| rpl37 | 0.068 | crabp2b | -0.033 |
| rcl1 | 0.068 | tmsb | -0.033 |
| gins2 | 0.067 | elavl4 | -0.033 |
| atic | 0.067 | rtn1b | -0.033 |
| rps17 | 0.067 | COX7A2 | -0.033 |
| rnaseh2a | 0.067 | tuba1c | -0.032 |
| rplp0 | 0.066 | nova1 | -0.032 |
| nifk | 0.066 | tmeff1b | -0.032 |