ring finger and WD repeat domain 3
ZFIN 
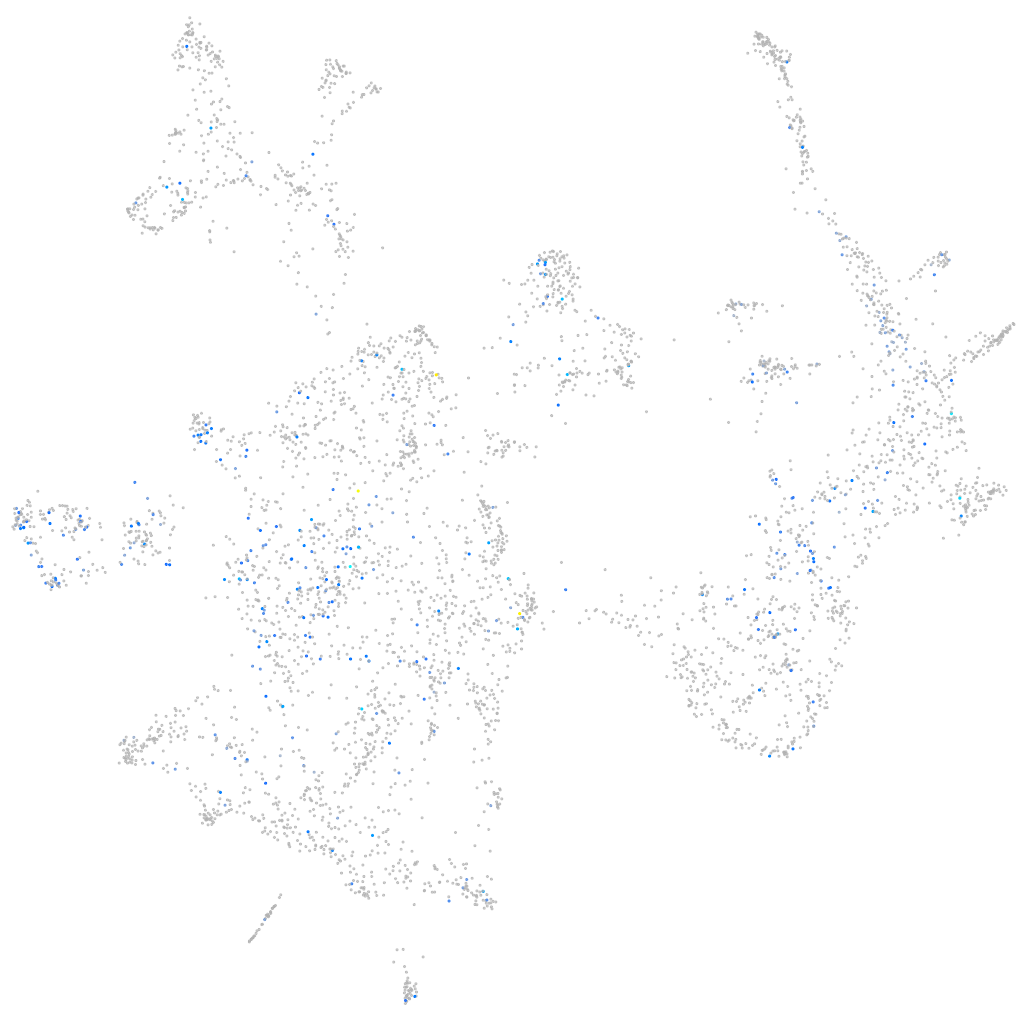
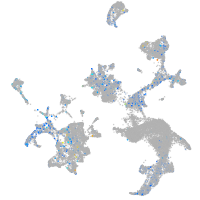
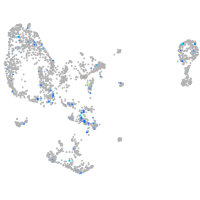
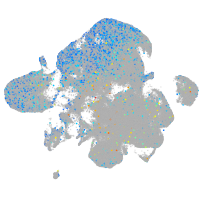
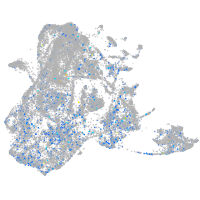
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.205 | pvalb2 | -0.077 |
| nasp | 0.199 | tob1b | -0.073 |
| chaf1a | 0.187 | ccni | -0.066 |
| rpa2 | 0.183 | b2ml | -0.066 |
| dhfr | 0.183 | glula | -0.061 |
| mibp | 0.182 | h1f0 | -0.060 |
| chtf18 | 0.180 | si:ch211-196l7.4 | -0.060 |
| dut | 0.179 | COX3 | -0.059 |
| rrm1 | 0.176 | tmem59 | -0.059 |
| rrm2 | 0.176 | zgc:158463 | -0.059 |
| prim2 | 0.171 | cebpd | -0.059 |
| LOC108179355 | 0.170 | pvalb1 | -0.058 |
| stmn1a | 0.169 | anxa5a | -0.058 |
| cnbpa | 0.166 | calm1b | -0.057 |
| tubb2b | 0.164 | CELA1 (1 of many) | -0.056 |
| cbx3a | 0.161 | dnajc5b | -0.055 |
| si:ch211-288g17.3 | 0.161 | calml4a | -0.055 |
| ccna2 | 0.158 | osbpl1a | -0.055 |
| hmgb2a | 0.158 | gpx2 | -0.054 |
| ranbp1 | 0.157 | si:ch73-15n15.3 | -0.054 |
| seta | 0.157 | enosf1 | -0.054 |
| asf1ba | 0.157 | hagh | -0.053 |
| hmga1a | 0.156 | kitlga | -0.053 |
| rnaseh2a | 0.155 | mylz3 | -0.053 |
| rpa3 | 0.155 | atp2b1a | -0.053 |
| cdca5 | 0.155 | eml2 | -0.052 |
| mcm7 | 0.155 | thbs1b | -0.052 |
| nutf2l | 0.154 | CR925719.1 | -0.052 |
| ncapg | 0.154 | gapdhs | -0.052 |
| dek | 0.154 | otofb | -0.052 |
| mad2l1 | 0.153 | sik1 | -0.052 |
| atad5a | 0.153 | apoa1b | -0.051 |
| banf1 | 0.153 | pvalb8 | -0.051 |
| zgc:110425 | 0.152 | hbegfa | -0.051 |
| si:dkey-6i22.5 | 0.152 | appb | -0.051 |