ring finger and WD repeat domain 3
ZFIN 
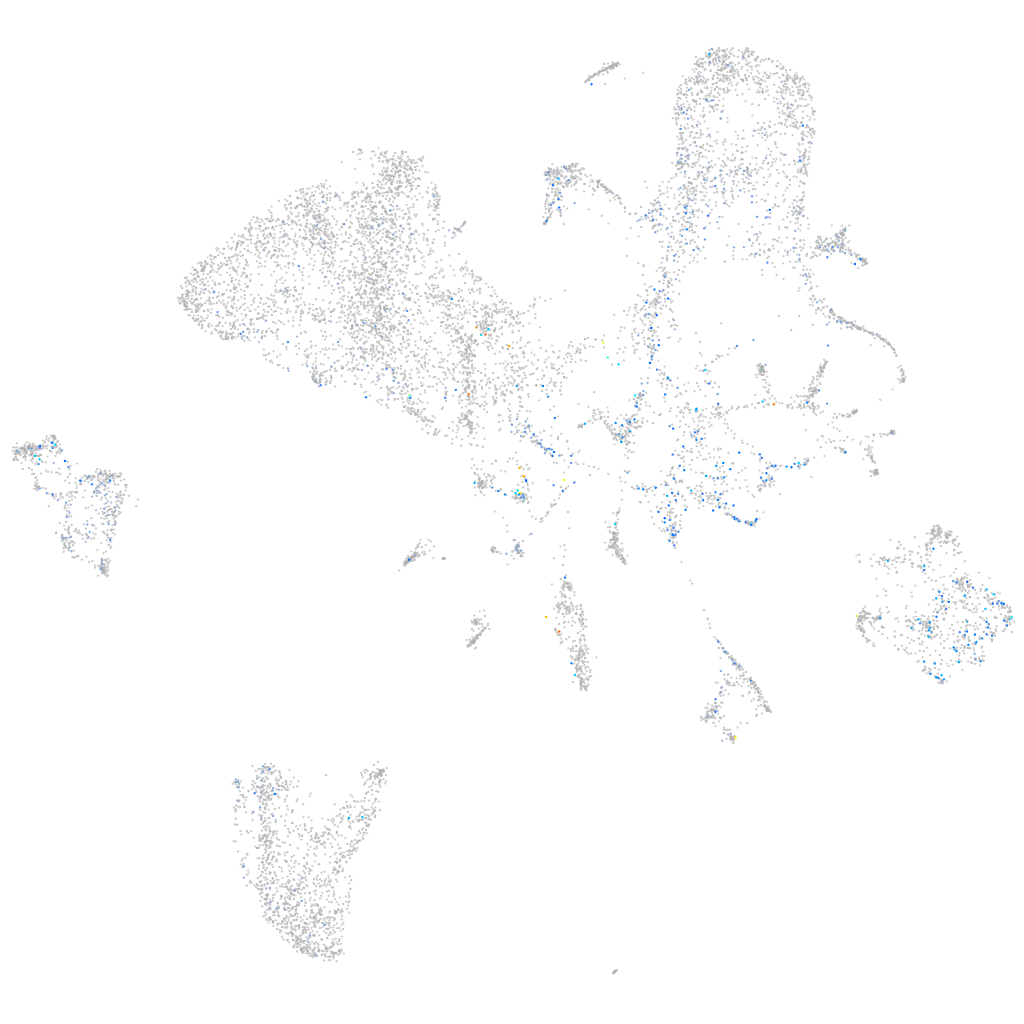
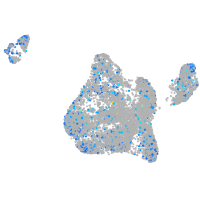
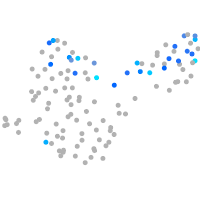
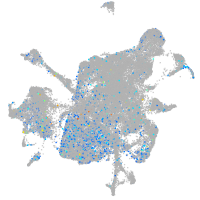
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.195 | gamt | -0.123 |
| sumo3b | 0.182 | gatm | -0.117 |
| tuba8l4 | 0.175 | pnp4b | -0.113 |
| nasp | 0.175 | gapdh | -0.108 |
| khdrbs1a | 0.175 | ahcy | -0.106 |
| h2afva | 0.173 | nupr1b | -0.103 |
| hmga1a | 0.173 | gpx4a | -0.101 |
| hmgb2a | 0.172 | bhmt | -0.097 |
| stmn1a | 0.172 | gc | -0.097 |
| hmgb2b | 0.171 | abat | -0.097 |
| setb | 0.171 | agxtb | -0.096 |
| si:ch211-222l21.1 | 0.171 | gnmt | -0.095 |
| hnrnpaba | 0.171 | pklr | -0.095 |
| rpa3 | 0.170 | apoa2 | -0.095 |
| cbx3a | 0.170 | zgc:92744 | -0.095 |
| tubb2b | 0.170 | fbp1b | -0.095 |
| hnrnpabb | 0.169 | mat1a | -0.095 |
| anp32b | 0.166 | fetub | -0.095 |
| dut | 0.165 | ces2 | -0.095 |
| h2afvb | 0.165 | scp2a | -0.094 |
| rbbp4 | 0.165 | g6pca.2 | -0.094 |
| hdac1 | 0.163 | apoa4b.1 | -0.093 |
| lig1 | 0.162 | ttr | -0.093 |
| syncrip | 0.162 | apoa1b | -0.093 |
| cirbpa | 0.162 | hao1 | -0.092 |
| chaf1a | 0.161 | etnppl | -0.092 |
| rpa2 | 0.160 | mgst1.2 | -0.092 |
| snrpd1 | 0.160 | apobb.1 | -0.091 |
| srsf3b | 0.160 | si:dkey-86l18.10 | -0.091 |
| hnrnpa0b | 0.160 | cyp2ad2 | -0.091 |
| cks1b | 0.159 | shbg | -0.091 |
| rrm1 | 0.159 | tdo2a | -0.091 |
| slbp | 0.159 | rbp4 | -0.090 |
| mis12 | 0.158 | rbp2b | -0.090 |
| si:ch211-288g17.3 | 0.158 | c9 | -0.090 |