RNA binding motif protein 7
ZFIN 
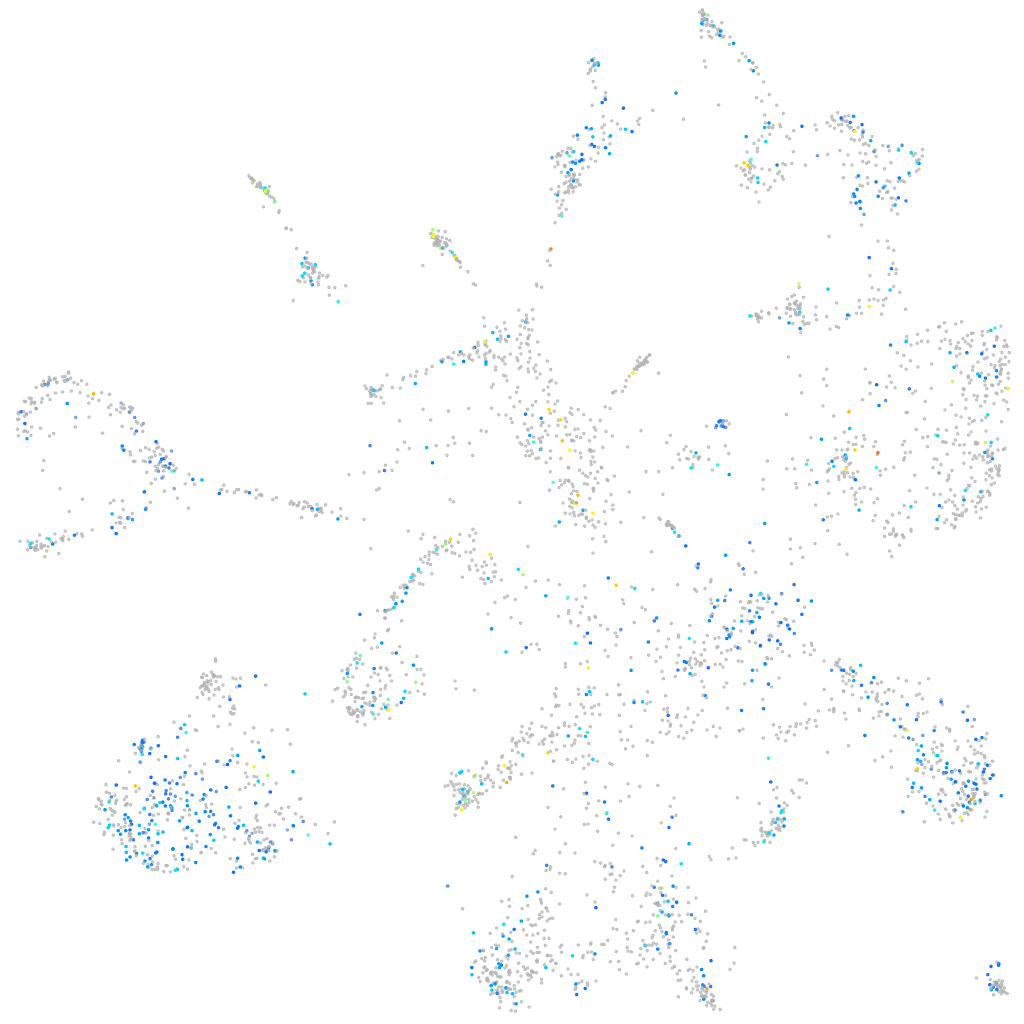
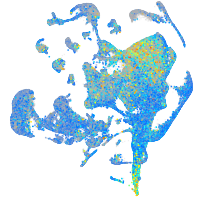
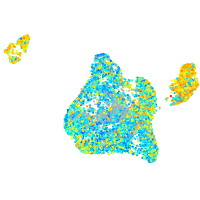
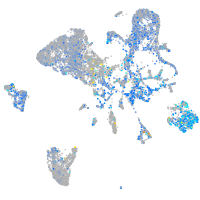
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.140 | amotl2a | -0.052 |
| khdrbs1a | 0.139 | kank2 | -0.052 |
| hnrnpabb | 0.133 | cavin2b | -0.051 |
| snrpd2 | 0.130 | pdk2b | -0.050 |
| srsf3b | 0.129 | c1qtnf12 | -0.049 |
| hnrnpa0l | 0.128 | si:dkey-51e6.1 | -0.047 |
| tubb5 | 0.127 | zgc:158463 | -0.045 |
| hmgn7 | 0.126 | rpl29 | -0.045 |
| hnrnpa0b | 0.124 | cav2 | -0.045 |
| tmeff1b | 0.123 | sik3 | -0.042 |
| marcksb | 0.123 | cxcl8b.3 | -0.042 |
| si:ch211-137a8.4 | 0.120 | LOC101883361 | -0.042 |
| h2afva | 0.119 | pmp22b | -0.042 |
| cnbpa | 0.119 | ela2 | -0.042 |
| hmgb1b | 0.119 | CABZ01072614.1 | -0.041 |
| atrx | 0.119 | hbae1.3 | -0.041 |
| tuba1c | 0.118 | pthlha | -0.040 |
| sumo3a | 0.118 | klf9 | -0.040 |
| bckdhb | 0.116 | aqp8a.1 | -0.040 |
| calm3b | 0.115 | krt8 | -0.039 |
| nrn1la | 0.114 | krt91 | -0.039 |
| syncrip | 0.113 | rhag | -0.039 |
| snrpd1 | 0.113 | CABZ01070631.1 | -0.039 |
| psma2 | 0.113 | slc2a1b | -0.038 |
| ywhabb | 0.112 | cdc42ep1a | -0.038 |
| tubb2b | 0.111 | rps29 | -0.038 |
| rtn1b | 0.111 | col6a3 | -0.038 |
| CR759927.3 | 0.110 | slc16a3 | -0.038 |
| dcaf7 | 0.110 | paplna | -0.037 |
| hmgb3a | 0.109 | CU856520.1 | -0.037 |
| setb | 0.108 | cavin1b | -0.037 |
| tubb4b | 0.108 | LOC110439372 | -0.037 |
| cct4 | 0.108 | lpin1 | -0.037 |
| anp32b | 0.107 | tmem100a | -0.036 |
| ubc | 0.107 | stat5a | -0.036 |