RNA binding motif protein 7
ZFIN 
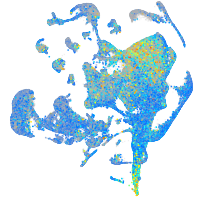
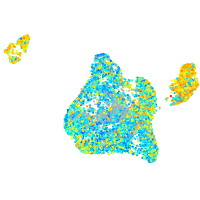
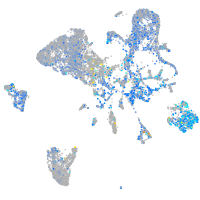
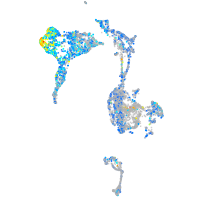
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.130 | atp1a1b | -0.073 |
| hnrnpa0l | 0.126 | ptn | -0.069 |
| h3f3d | 0.121 | glula | -0.060 |
| khdrbs1a | 0.111 | cx43 | -0.058 |
| cirbpb | 0.106 | slc4a4a | -0.057 |
| hnrnpabb | 0.105 | fabp7a | -0.053 |
| hnrnpa0b | 0.105 | si:ch211-66e2.5 | -0.052 |
| tubb2b | 0.102 | mt2 | -0.051 |
| ptmab | 0.101 | cebpd | -0.051 |
| si:ch211-288g17.3 | 0.100 | slc1a2b | -0.050 |
| cct5 | 0.100 | sparc | -0.050 |
| sumo3a | 0.098 | gpr37l1b | -0.049 |
| hmgn6 | 0.098 | zgc:158463 | -0.049 |
| h3f3a | 0.097 | slc3a2a | -0.049 |
| marcksb | 0.096 | cyp2ad3 | -0.049 |
| h2afvb | 0.096 | cox4i2 | -0.047 |
| hsp90ab1 | 0.095 | qki2 | -0.047 |
| ilf2 | 0.094 | mdkb | -0.047 |
| smarce1 | 0.093 | atp1b4 | -0.046 |
| ddx39ab | 0.092 | zgc:153704 | -0.045 |
| srsf3b | 0.092 | efhd1 | -0.045 |
| hdac1 | 0.091 | apoa1b | -0.045 |
| erh | 0.090 | slit2 | -0.044 |
| nono | 0.090 | cldn19 | -0.044 |
| bud31 | 0.090 | clstn2 | -0.043 |
| ybx1 | 0.089 | ptgdsb.2 | -0.043 |
| si:ch1073-429i10.3.1 | 0.088 | actc1b | -0.043 |
| cct3 | 0.087 | s100b | -0.043 |
| cct2 | 0.087 | prss1 | -0.043 |
| hmga1a | 0.087 | pygmb | -0.042 |
| syncrip | 0.087 | ppap2d | -0.042 |
| psmb7 | 0.086 | igfbp7 | -0.042 |
| hmgb1b | 0.086 | prss59.2 | -0.041 |
| sumo2b | 0.086 | mlc1 | -0.041 |
| h2afva | 0.086 | acbd7 | -0.041 |