ribonucleic acid export 1
ZFIN 
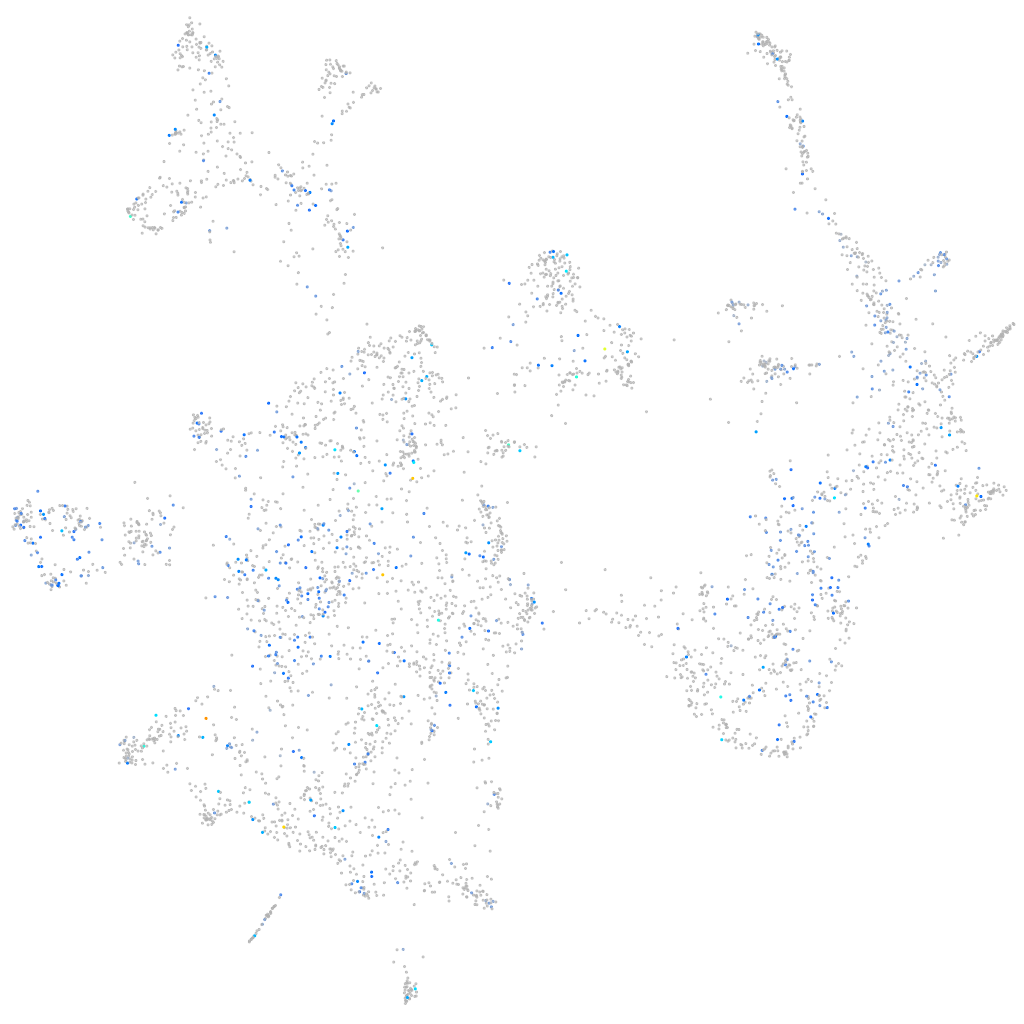
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR954285.1 | 0.170 | pvalb1 | -0.079 |
| BX465838.1 | 0.168 | ccni | -0.066 |
| dut | 0.159 | sik1 | -0.064 |
| LO018550.2 | 0.153 | apoa2 | -0.062 |
| ftr04 | 0.152 | nupr1a | -0.060 |
| seta | 0.147 | gsto2 | -0.057 |
| hmgb2b | 0.144 | b2ml | -0.057 |
| pcna | 0.144 | tob1b | -0.056 |
| si:ch211-288g17.3 | 0.143 | cd164l2 | -0.055 |
| tubb2b | 0.143 | pvalb2 | -0.053 |
| rpa3 | 0.142 | actc1b | -0.053 |
| ranbp1 | 0.142 | cebpd | -0.052 |
| hmga1a | 0.140 | abca1a | -0.052 |
| hnrnpaba | 0.139 | cyp3c1 | -0.052 |
| taf15 | 0.137 | vsig10l2 | -0.051 |
| ddx39ab | 0.136 | tmem59 | -0.051 |
| rrm1 | 0.135 | eno1a | -0.050 |
| stmn1a | 0.134 | mcl1a | -0.050 |
| snu13b | 0.134 | acta1b | -0.049 |
| FO834800.2 | 0.133 | ddit3 | -0.049 |
| rbbp4 | 0.132 | apba1a | -0.049 |
| sae1 | 0.132 | gapdhs | -0.048 |
| khdrbs1a | 0.131 | trim36 | -0.048 |
| hmgn2 | 0.131 | tob1a | -0.048 |
| snrpd1 | 0.130 | ttn.1 | -0.048 |
| rrm2 | 0.130 | si:ch211-196l7.4 | -0.048 |
| nasp | 0.130 | necap1 | -0.048 |
| suv39h1b | 0.129 | ctsf | -0.047 |
| snrpf | 0.129 | CR383676.1 | -0.047 |
| si:ch211-222l21.1 | 0.128 | socs3a | -0.047 |
| hmgb2a | 0.128 | CR925719.1 | -0.047 |
| nop58 | 0.128 | gpx2 | -0.047 |
| pdap1a | 0.128 | si:dkey-11c5.11 | -0.047 |
| hnrnpabb | 0.127 | gabarapl2 | -0.046 |
| banf1 | 0.127 | gfi1aa | -0.046 |