"RAB6B, member RAS oncogene family b"
ZFIN 
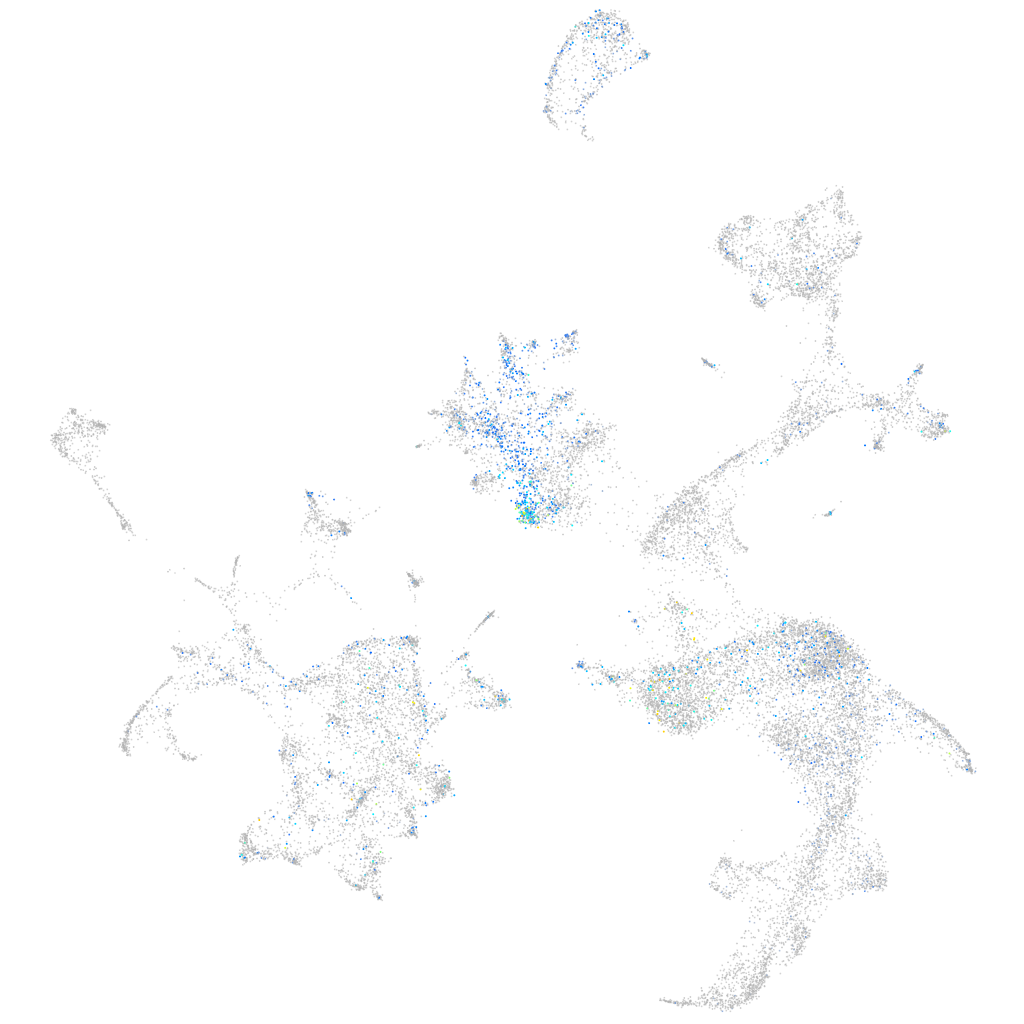
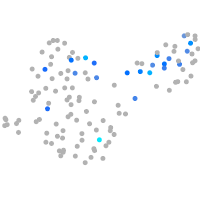
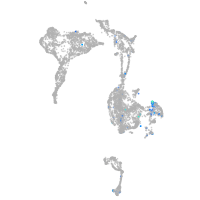
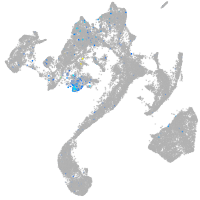
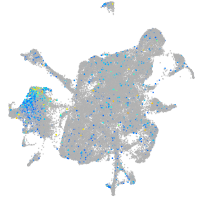
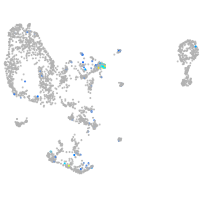
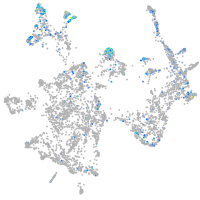
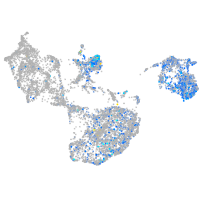
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn2a | 0.289 | rps12 | -0.079 |
| sncb | 0.275 | hbbe3 | -0.077 |
| zgc:65894 | 0.271 | urod | -0.072 |
| atp6v0cb | 0.270 | znfl2a | -0.069 |
| ywhag2 | 0.269 | hmbsa | -0.069 |
| gng3 | 0.266 | npm1a | -0.068 |
| stxbp1a | 0.264 | pabpc1a | -0.066 |
| vamp2 | 0.260 | hmbsb | -0.065 |
| snap25a | 0.259 | blf | -0.065 |
| gap43 | 0.258 | dkc1 | -0.065 |
| elavl4 | 0.250 | serbp1a | -0.063 |
| mllt11 | 0.249 | alad | -0.063 |
| sv2a | 0.247 | rplp2l | -0.063 |
| rtn1b | 0.243 | cpox | -0.063 |
| tmsb2 | 0.241 | rps6 | -0.062 |
| tuba1c | 0.241 | nop58 | -0.062 |
| atpv0e2 | 0.240 | nutf2l | -0.061 |
| stmn1b | 0.239 | fbl | -0.061 |
| map1aa | 0.238 | anp32b | -0.061 |
| sncgb | 0.234 | rps29 | -0.059 |
| stx1b | 0.230 | nhp2 | -0.059 |
| syn2a | 0.229 | cldng | -0.059 |
| eno2 | 0.227 | rpl29 | -0.059 |
| scg2b | 0.227 | nop2 | -0.058 |
| si:dkeyp-75h12.5 | 0.226 | pcna | -0.058 |
| cnrip1a | 0.225 | rps26l | -0.057 |
| necap1 | 0.225 | abcf1 | -0.056 |
| zgc:153426 | 0.224 | banf1 | -0.056 |
| atp6v1b2 | 0.223 | snu13b | -0.056 |
| rnasekb | 0.222 | si:dkey-261j4.4 | -0.055 |
| syt11a | 0.220 | rpl12 | -0.055 |
| slc6a1a | 0.220 | chaf1a | -0.055 |
| cspg5a | 0.220 | rrs1 | -0.055 |
| syt1a | 0.217 | gnl3 | -0.055 |
| gpm6aa | 0.216 | mcm2 | -0.054 |