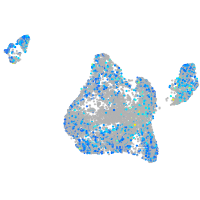
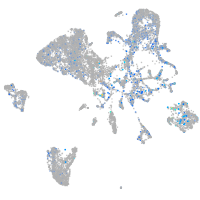
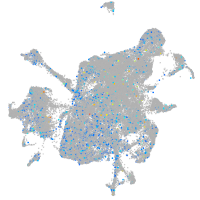
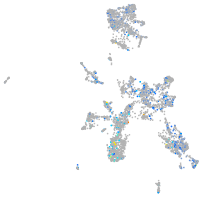
PC4 and SFRS1 interacting protein 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| id1 | 0.066 | gpm6ab | -0.054 |
| hmgb2a | 0.064 | stmn1b | -0.051 |
| mki67 | 0.063 | rtn1a | -0.049 |
| ranbp1 | 0.062 | ptmaa | -0.048 |
| eif5a2 | 0.060 | gpm6aa | -0.045 |
| sox3 | 0.059 | atp6v0cb | -0.044 |
| tuba8l4 | 0.058 | pvalb1 | -0.043 |
| cldn5a | 0.057 | gng3 | -0.043 |
| hmga1a | 0.057 | rtn1b | -0.043 |
| ptges3b | 0.056 | pvalb2 | -0.042 |
| snrpf | 0.056 | actc1b | -0.042 |
| tpx2 | 0.056 | ckbb | -0.041 |
| top2a | 0.055 | sncb | -0.041 |
| banf1 | 0.055 | elavl4 | -0.040 |
| tuba8l | 0.055 | aplp1 | -0.040 |
| XLOC-003690 | 0.054 | rnasekb | -0.038 |
| ran | 0.054 | si:dkeyp-75h12.5 | -0.038 |
| sox19a | 0.054 | stx1b | -0.036 |
| mad2l1 | 0.054 | nsg2 | -0.035 |
| cbx3a | 0.054 | syt1a | -0.035 |
| anp32b | 0.053 | vamp2 | -0.035 |
| gfap | 0.053 | elavl3 | -0.035 |
| plk1 | 0.053 | nrxn1a | -0.034 |
| rpl5a | 0.053 | stxbp1a | -0.034 |
| msna | 0.053 | ywhag2 | -0.033 |
| hmgb2b | 0.053 | snap25a | -0.033 |
| chaf1a | 0.053 | id4 | -0.032 |
| snrpe | 0.053 | gabrb4 | -0.032 |
| msi1 | 0.053 | olfm1b | -0.032 |
| selenoh | 0.053 | tmem59l | -0.032 |
| serbp1a | 0.053 | myt1la | -0.032 |
| rrm1 | 0.052 | map1aa | -0.032 |
| CABZ01075068.1 | 0.052 | tmsb2 | -0.032 |
| XLOC-003689 | 0.052 | sox4a | -0.031 |
| ccna2 | 0.052 | hbae3 | -0.031 |