proline rich 14
ZFIN 
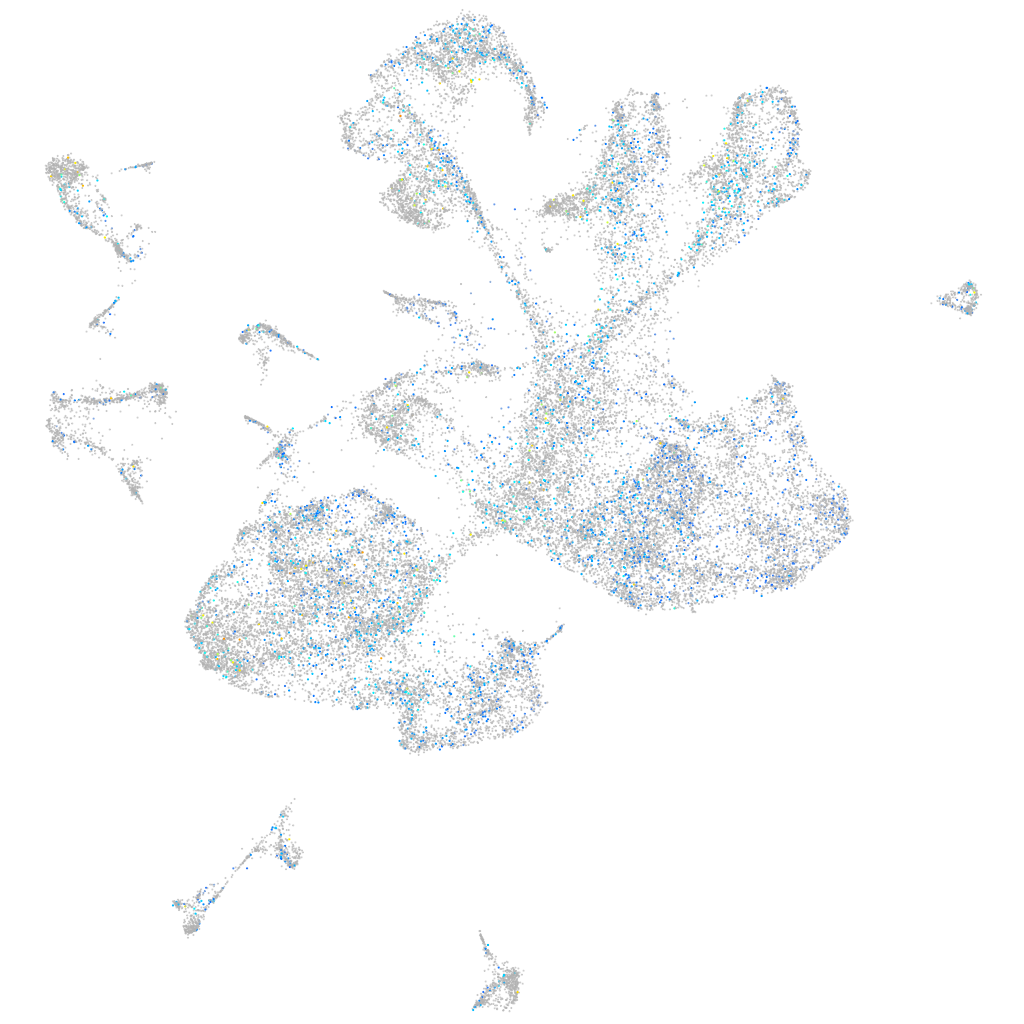
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chd4a | 0.066 | ldlrad2 | -0.035 |
| atrx | 0.063 | oc90 | -0.031 |
| hmgb1b | 0.062 | apela | -0.031 |
| hmgb3a | 0.058 | CABZ01075068.1 | -0.028 |
| hmgb1a | 0.057 | cdx4 | -0.028 |
| tmeff1b | 0.056 | apcdd1l | -0.026 |
| tmpob | 0.056 | apoa2 | -0.026 |
| hnrnpa0a | 0.053 | eif4ebp3l | -0.026 |
| tubb2b | 0.053 | gstp1 | -0.026 |
| si:ch211-288g17.3 | 0.053 | XLOC-042222 | -0.025 |
| CU634008.1 | 0.053 | sparc | -0.024 |
| tuba1c | 0.053 | FQ323156.1 | -0.024 |
| zswim5 | 0.051 | hspb1 | -0.024 |
| hnrnpaba | 0.051 | zgc:112437 | -0.024 |
| seta | 0.050 | cyp2ad3 | -0.024 |
| si:dkey-276j7.1 | 0.050 | BX001014.2 | -0.024 |
| si:ch73-46j18.5 | 0.050 | XLOC-001964 | -0.024 |
| baz1b | 0.050 | mt2 | -0.023 |
| ptmab | 0.050 | mcl1b | -0.023 |
| nfil3-5 | 0.050 | meig1 | -0.023 |
| myt1b | 0.049 | mfap2 | -0.023 |
| nova2 | 0.049 | glula | -0.023 |
| zfhx3 | 0.049 | slc4a4a | -0.023 |
| marcksb | 0.048 | s100b | -0.023 |
| top2b | 0.048 | gpx4a | -0.022 |
| FO082781.1 | 0.048 | col8a1a | -0.022 |
| nsg2 | 0.048 | wnt11r | -0.022 |
| nucks1a | 0.047 | tbata | -0.022 |
| cirbpb | 0.047 | col9a3 | -0.021 |
| hnrnpa1a | 0.047 | aplnr2 | -0.021 |
| smarcd1 | 0.047 | clu | -0.021 |
| h3f3a | 0.046 | fstl1a | -0.021 |
| ankrd12 | 0.046 | lamb1a | -0.021 |
| tmsb | 0.046 | rsl1d1 | -0.021 |
| cfl1 | 0.046 | zgc:56699 | -0.021 |