POU class 6 homeobox 2
ZFIN 
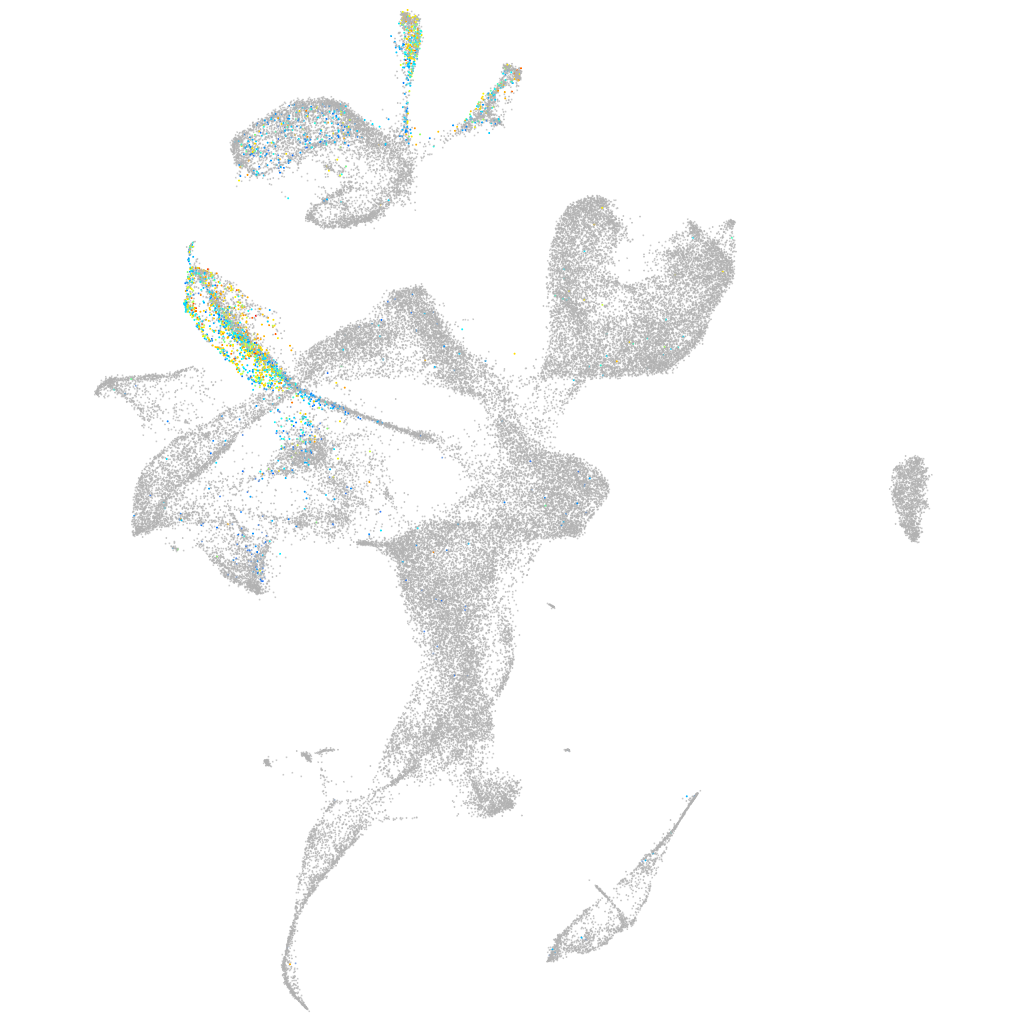
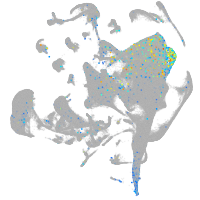
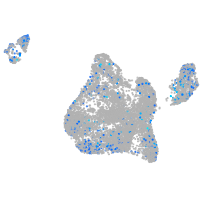
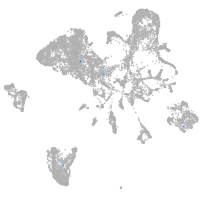
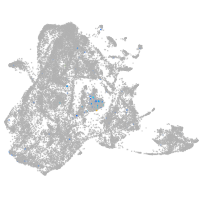
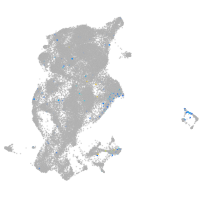
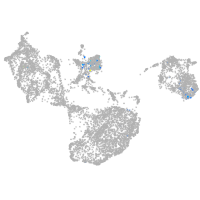
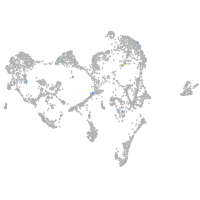
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| isl2b | 0.440 | hmgb2a | -0.229 |
| inab | 0.436 | mdka | -0.119 |
| rbpms2a | 0.434 | msi1 | -0.117 |
| rbpms2b | 0.418 | pcna | -0.115 |
| islr2 | 0.406 | hmga1a | -0.115 |
| zfhx3 | 0.392 | stmn1a | -0.111 |
| xpr1a | 0.352 | msna | -0.110 |
| gap43 | 0.339 | fabp7a | -0.110 |
| stmn2b | 0.334 | anp32e | -0.107 |
| tmsb | 0.329 | dut | -0.106 |
| BX247868.1 | 0.328 | nutf2l | -0.106 |
| zgc:158291 | 0.327 | neurod4 | -0.105 |
| ebf3a | 0.325 | rrm1 | -0.105 |
| stmn2a | 0.315 | syt5b | -0.104 |
| elavl3 | 0.311 | ccnd1 | -0.104 |
| id4 | 0.306 | crx | -0.103 |
| rtn1b | 0.300 | mki67 | -0.102 |
| irx4a | 0.298 | ahcy | -0.102 |
| nrn1a | 0.297 | selenoh | -0.101 |
| cntn2 | 0.296 | otx5 | -0.100 |
| gnao1a | 0.283 | dek | -0.100 |
| map7d2b | 0.281 | tuba8l | -0.099 |
| stmn1b | 0.279 | chaf1a | -0.099 |
| si:dkey-280e21.3 | 0.279 | lbr | -0.099 |
| cplx2l | 0.276 | mcm7 | -0.099 |
| pou4f1 | 0.275 | her15.1 | -0.096 |
| mllt11 | 0.273 | banf1 | -0.096 |
| si:ch211-129p13.1 | 0.271 | eef1da | -0.095 |
| gng3 | 0.270 | rpa3 | -0.095 |
| pou4f2 | 0.270 | ccna2 | -0.095 |
| stmn4l | 0.268 | anp32b | -0.094 |
| zgc:101840 | 0.266 | fen1 | -0.094 |
| cplx2 | 0.264 | COX7A2 (1 of many) | -0.091 |
| eno2 | 0.262 | id1 | -0.091 |
| shisal1a | 0.258 | si:ch211-156b7.4 | -0.090 |