POU class 6 homeobox 2
ZFIN 
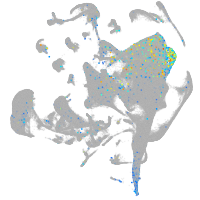
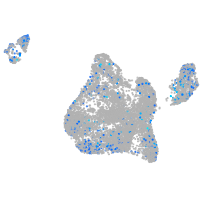
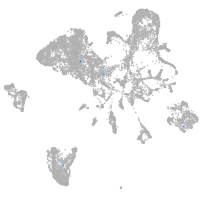
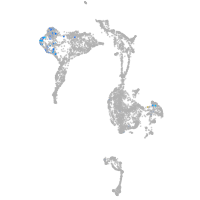
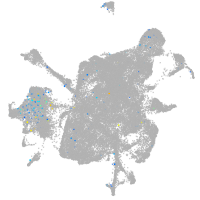
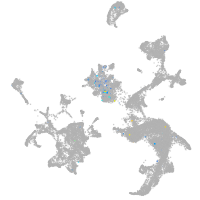
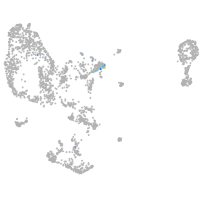
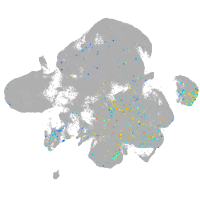
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| isl2a | 0.156 | XLOC-003690 | -0.097 |
| cdh5 | 0.153 | her15.1 | -0.094 |
| onecut1 | 0.148 | id1 | -0.088 |
| elavl4 | 0.143 | XLOC-003689 | -0.082 |
| prph | 0.141 | hmgb2a | -0.081 |
| isl2b | 0.140 | mdka | -0.080 |
| inab | 0.140 | cldn5a | -0.079 |
| islr2 | 0.138 | sox3 | -0.074 |
| tac1 | 0.138 | btg2 | -0.072 |
| stmn1b | 0.137 | tspan7 | -0.072 |
| gng3 | 0.133 | sdc4 | -0.070 |
| nsmfb | 0.131 | notch3 | -0.069 |
| rtn1b | 0.127 | her4.2 | -0.068 |
| gap43 | 0.125 | COX7A2 (1 of many) | -0.066 |
| phox2a | 0.125 | rpl12 | -0.065 |
| isl1 | 0.125 | her12 | -0.065 |
| ppp1r14ba | 0.124 | XLOC-003692 | -0.065 |
| tmsb2 | 0.124 | dut | -0.064 |
| gabrb4 | 0.122 | vamp3 | -0.063 |
| sncb | 0.121 | msi1 | -0.063 |
| slit3 | 0.119 | msna | -0.063 |
| stmn2a | 0.119 | sparc | -0.063 |
| lmo1 | 0.118 | sox19a | -0.063 |
| cplx2 | 0.118 | si:dkey-151g10.6 | -0.062 |
| tmsb | 0.118 | si:ch73-281n10.2 | -0.062 |
| slc5a7a | 0.118 | sb:cb81 | -0.061 |
| tuba2 | 0.118 | rplp1 | -0.060 |
| syt2a | 0.117 | her4.1 | -0.060 |
| cxxc4 | 0.117 | hmgb2b | -0.059 |
| gpm6ab | 0.117 | rps2 | -0.059 |
| chata | 0.115 | pcna | -0.059 |
| id4 | 0.114 | rpl39 | -0.058 |
| golga7ba | 0.113 | rps7 | -0.058 |
| slc18a3a | 0.112 | abhd6a | -0.058 |
| stmn4l | 0.111 | eef1a1l1 | -0.058 |