polycystic kidney disease 1a
ZFIN 
Other cell groups


all cells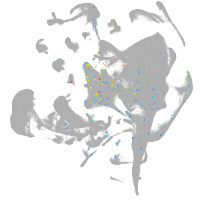 |
blastomeres |
PGCs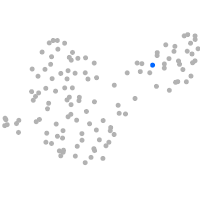 |
endoderm |
axial mesoderm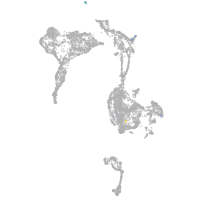 |
muscle 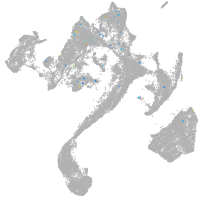 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 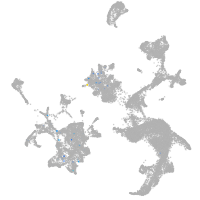 |
pronephros 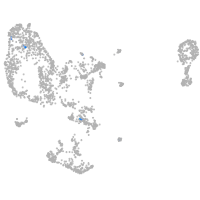 |
neural |
spinal cord / glia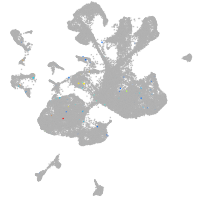 |
eye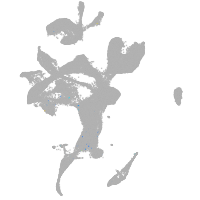 |
taste / olfactory |
otic / lateral line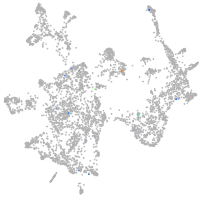 |
epidermis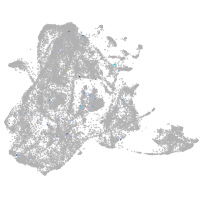 |
periderm |
fin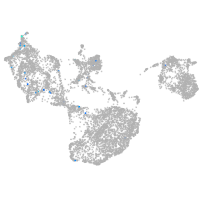 |
ionocytes / mucous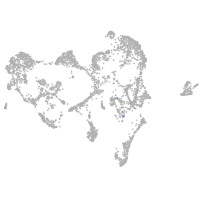 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX927308.2 | 0.079 | rtn1a | -0.028 |
| cdcp2 | 0.072 | elavl3 | -0.025 |
| fbln5 | 0.072 | gng3 | -0.024 |
| CABZ01113373.1 | 0.059 | stmn1b | -0.024 |
| BX928743.1 | 0.055 | sncb | -0.023 |
| CT009596.1 | 0.055 | ckbb | -0.021 |
| LOC101884267 | 0.053 | tuba1c | -0.021 |
| golim4a | 0.052 | zc4h2 | -0.021 |
| amfrb | 0.051 | rnasekb | -0.021 |
| FP102018.1 | 0.050 | rtn1b | -0.021 |
| ppfibp1a | 0.049 | atp6v0cb | -0.020 |
| ryr2b | 0.048 | fez1 | -0.020 |
| CABZ01071326.1 | 0.048 | gpm6aa | -0.020 |
| BX088712.1 | 0.044 | tmsb | -0.020 |
| rfesd | 0.043 | vamp2 | -0.019 |
| ehd2b | 0.043 | gpm6ab | -0.019 |
| BX890600.1 | 0.042 | stmn2a | -0.019 |
| mir214a | 0.042 | elavl4 | -0.019 |
| CR848683.1 | 0.041 | gap43 | -0.019 |
| CR376741.2 | 0.040 | atp6v1g1 | -0.018 |
| sh3tc2 | 0.040 | mllt11 | -0.018 |
| LOC101882228 | 0.040 | si:dkeyp-75h12.5 | -0.018 |
| mylka | 0.040 | gng2 | -0.018 |
| CT009487.2 | 0.039 | zgc:65894 | -0.017 |
| cd37 | 0.038 | ywhag2 | -0.017 |
| ror1 | 0.037 | calm1a | -0.017 |
| thbs1b | 0.036 | gdi1 | -0.017 |
| il16 | 0.035 | si:dkey-276j7.1 | -0.017 |
| LOC101882122 | 0.035 | tubb5 | -0.017 |
| mir199-3 | 0.035 | id4 | -0.017 |
| si:dkey-261h17.1 | 0.035 | csdc2a | -0.017 |
| fstl1b | 0.034 | gnao1a | -0.016 |
| nkd2a | 0.034 | snap25a | -0.016 |
| ggh | 0.033 | nsg2 | -0.016 |
| tbx20 | 0.033 | tpi1b | -0.016 |




