PIN2 (TERF1) interacting telomerase inhibitor 1
ZFIN 
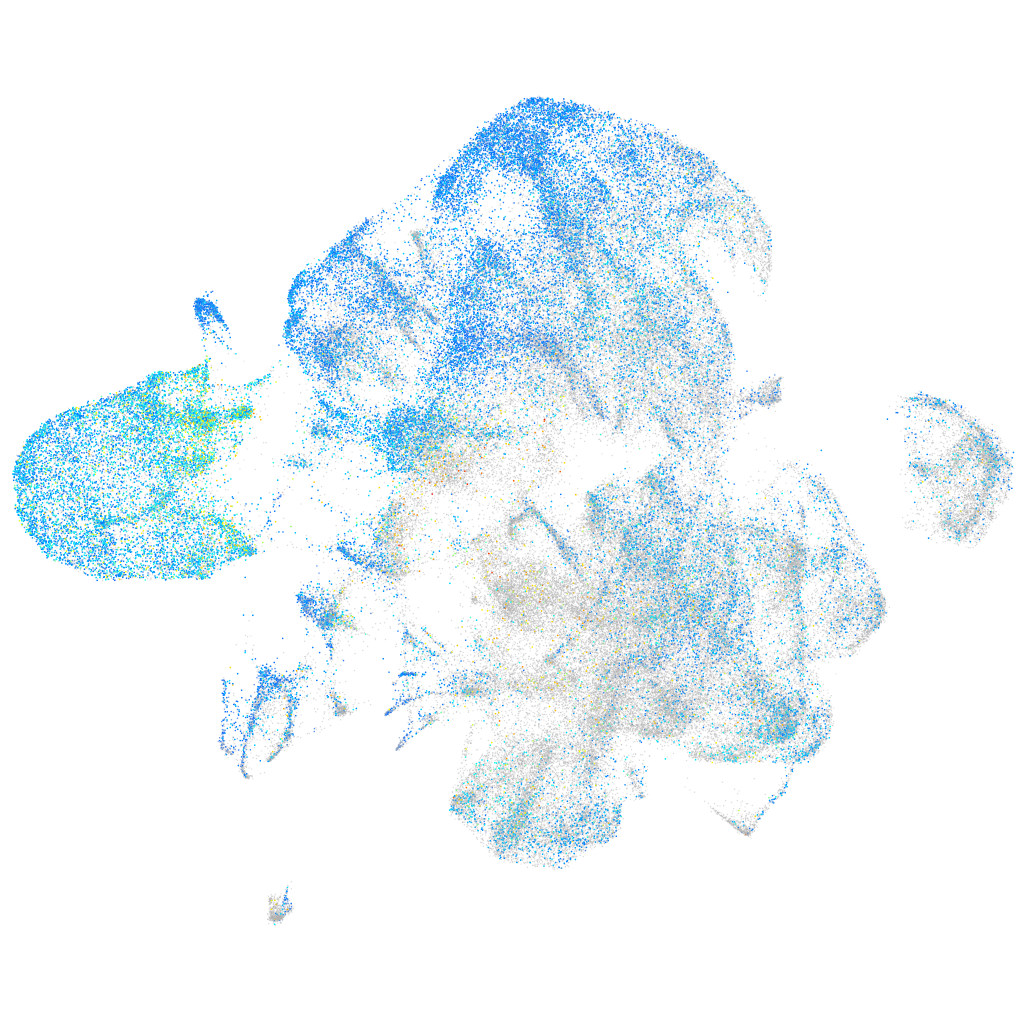
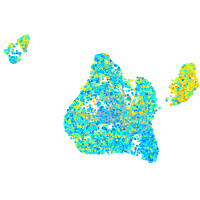
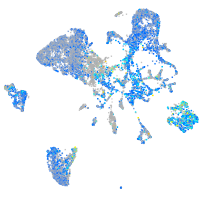
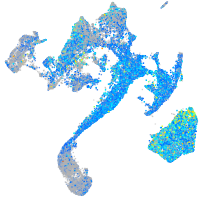
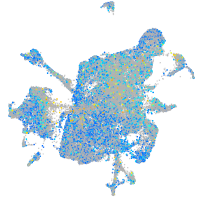
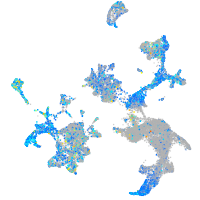
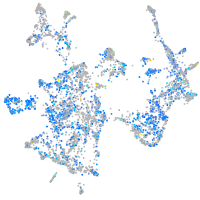
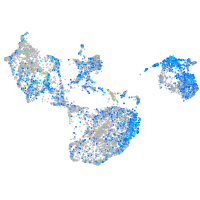
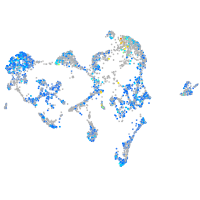
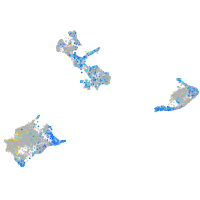
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.404 | ptmaa | -0.313 |
| nop58 | 0.393 | CR383676.1 | -0.276 |
| dkc1 | 0.384 | rtn1a | -0.273 |
| fbl | 0.374 | gpm6aa | -0.266 |
| nop56 | 0.361 | tuba1c | -0.261 |
| hspb1 | 0.359 | elavl3 | -0.255 |
| rsl1d1 | 0.354 | stmn1b | -0.249 |
| nop2 | 0.351 | rnasekb | -0.217 |
| snu13b | 0.351 | marcksl1a | -0.216 |
| ncl | 0.349 | gpm6ab | -0.216 |
| nhp2 | 0.326 | ckbb | -0.215 |
| ebna1bp2 | 0.325 | atp6v0cb | -0.213 |
| gnl3 | 0.322 | tmsb4x | -0.207 |
| nop10 | 0.317 | sncb | -0.202 |
| si:ch211-217k17.7 | 0.314 | COX3 | -0.197 |
| si:ch211-152c2.3 | 0.313 | gng3 | -0.192 |
| gar1 | 0.310 | ppdpfb | -0.192 |
| ddx18 | 0.308 | vamp2 | -0.192 |
| rrp1 | 0.307 | calm1a | -0.191 |
| bms1 | 0.304 | nova2 | -0.190 |
| pes | 0.303 | tmsb | -0.189 |
| lyar | 0.299 | pvalb1 | -0.188 |
| lig1 | 0.296 | elavl4 | -0.187 |
| mybbp1a | 0.296 | pvalb2 | -0.185 |
| aldob | 0.295 | actc1b | -0.181 |
| wdr43 | 0.293 | rtn1b | -0.181 |
| apoeb | 0.293 | rpl37 | -0.180 |
| pprc1 | 0.291 | si:dkeyp-75h12.5 | -0.180 |
| anp32b | 0.290 | h3f3a | -0.180 |
| zmp:0000000624 | 0.290 | atp6v1e1b | -0.179 |
| rrs1 | 0.288 | ywhag2 | -0.178 |
| rpl7l1 | 0.285 | ccni | -0.178 |
| mcm6 | 0.285 | gapdhs | -0.177 |
| eif4bb | 0.282 | tubb5 | -0.175 |
| eif5a2 | 0.282 | stx1b | -0.175 |