3-phosphoinositide dependent protein kinase 1b
ZFIN 
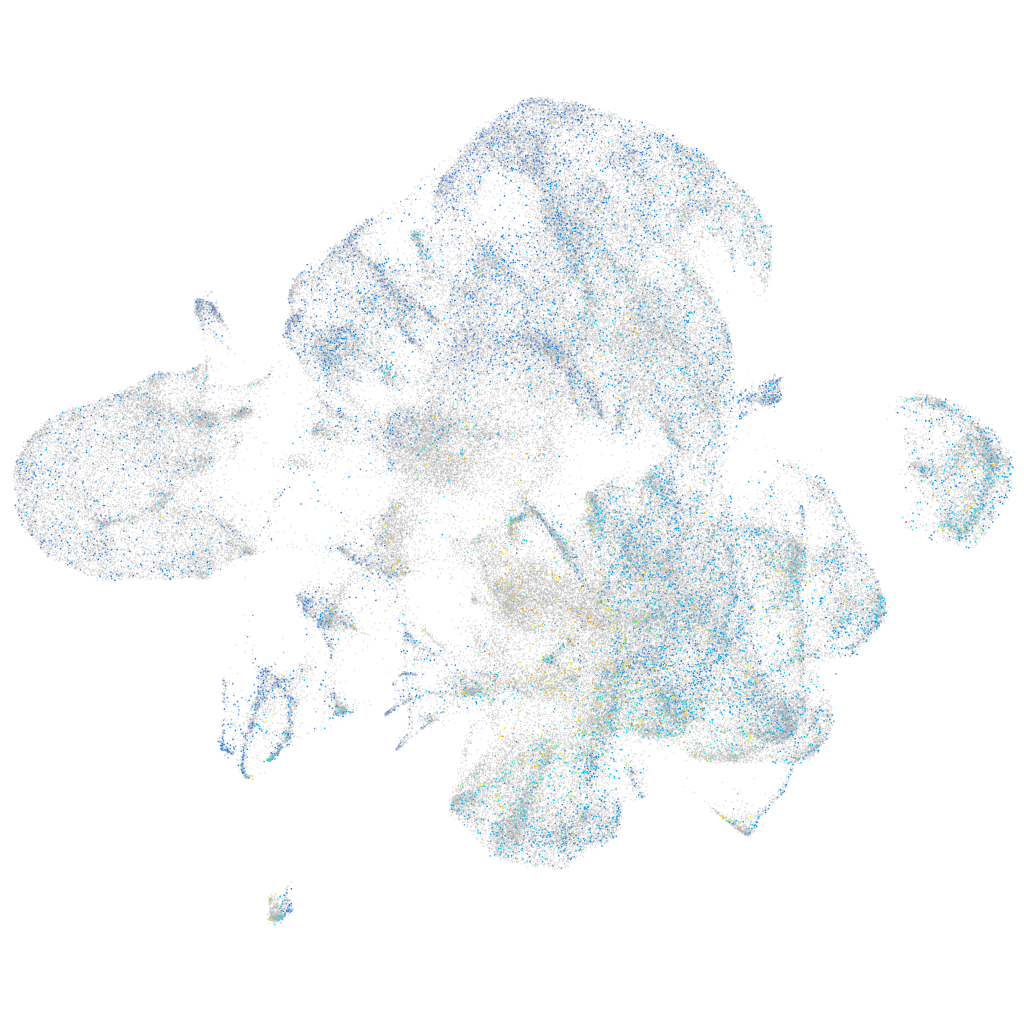
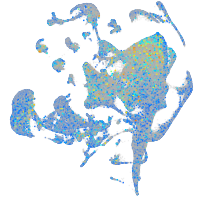
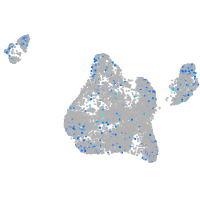
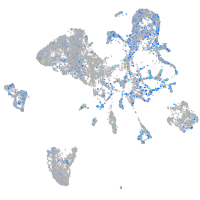
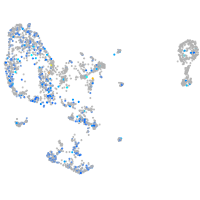
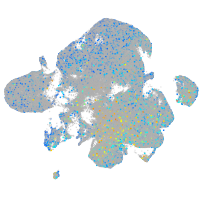
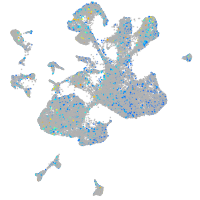
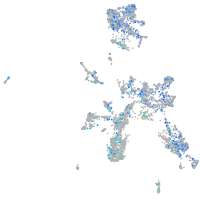
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ywhag2 | 0.094 | hmgb2a | -0.065 |
| stmn2a | 0.091 | hmga1a | -0.058 |
| calm1a | 0.091 | npm1a | -0.055 |
| gng3 | 0.090 | dkc1 | -0.054 |
| atp6v1e1b | 0.090 | nop58 | -0.051 |
| sncb | 0.090 | fbl | -0.050 |
| atp6v1g1 | 0.089 | hmgb2b | -0.050 |
| vamp2 | 0.088 | nop56 | -0.049 |
| snap25a | 0.086 | rsl1d1 | -0.049 |
| gapdhs | 0.086 | si:dkey-151g10.6 | -0.048 |
| zgc:65894 | 0.086 | rps12 | -0.048 |
| stxbp1a | 0.085 | rplp2l | -0.047 |
| calm2a | 0.085 | rplp1 | -0.047 |
| mllt11 | 0.084 | hspb1 | -0.047 |
| rnasekb | 0.082 | snu13b | -0.047 |
| calm1b | 0.082 | nop2 | -0.047 |
| tpi1b | 0.081 | rpl4 | -0.046 |
| atp6v0cb | 0.081 | ddx18 | -0.045 |
| tuba1c | 0.081 | gnl3 | -0.044 |
| necap1 | 0.080 | NC-002333.4 | -0.043 |
| elavl4 | 0.079 | cdca7b | -0.043 |
| stmn1b | 0.078 | nhp2 | -0.043 |
| gnao1a | 0.077 | id1 | -0.043 |
| gnb1a | 0.077 | rps28 | -0.043 |
| gap43 | 0.076 | rps6 | -0.043 |
| ckbb | 0.076 | cdca7a | -0.043 |
| stx1b | 0.075 | rps18 | -0.042 |
| ppdpfb | 0.075 | pcna | -0.042 |
| eno2 | 0.075 | ccnd1 | -0.042 |
| syn2a | 0.075 | nop10 | -0.041 |
| gabarapl2 | 0.074 | ube2c | -0.041 |
| hist2h2l | 0.074 | lig1 | -0.041 |
| atpv0e2 | 0.073 | eef1a1l1 | -0.041 |
| tmsb2 | 0.073 | mcm6 | -0.041 |
| calm3b | 0.073 | banf1 | -0.041 |