protocadherin 1 gamma 14
ZFIN 
Other cell groups


all cells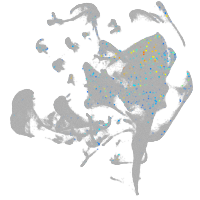 |
blastomeres |
PGCs |
endoderm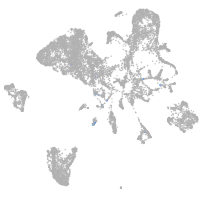 |
axial mesoderm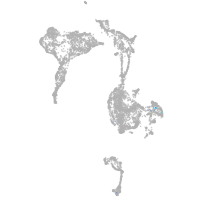 |
muscle 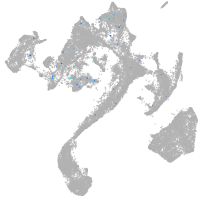 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia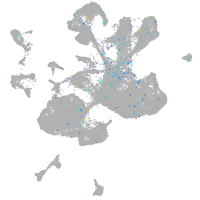 |
eye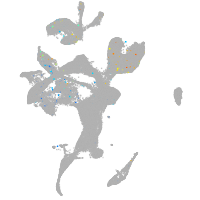 |
taste / olfactory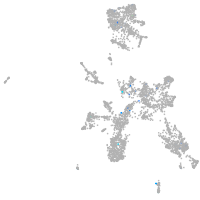 |
otic / lateral line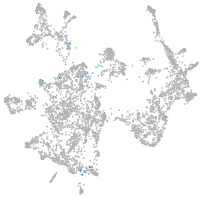 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ppp1r14c | 0.084 | pcna | -0.055 |
| gpm6aa | 0.081 | banf1 | -0.055 |
| scrt2 | 0.076 | msna | -0.054 |
| gpm6ab | 0.073 | stmn1a | -0.054 |
| nova2 | 0.073 | tuba8l4 | -0.053 |
| mdkb | 0.069 | rrm1 | -0.052 |
| nsg2 | 0.069 | lbr | -0.050 |
| rtn1a | 0.067 | dut | -0.050 |
| ndrg4 | 0.064 | tuba8l | -0.050 |
| gng13b | 0.063 | ccna2 | -0.048 |
| gnb3a | 0.062 | mcm7 | -0.048 |
| tmem178b | 0.062 | selenoh | -0.048 |
| hunk | 0.061 | ccnd1 | -0.048 |
| rorab | 0.061 | mki67 | -0.048 |
| zc4h2 | 0.060 | tma7 | -0.047 |
| tp53inp1 | 0.059 | chaf1a | -0.047 |
| zgc:100920 | 0.059 | nutf2l | -0.047 |
| prr18 | 0.058 | rpa3 | -0.046 |
| marcksl1a | 0.057 | anp32b | -0.046 |
| dscaml1 | 0.057 | dek | -0.046 |
| si:ch211-129p13.1 | 0.056 | fen1 | -0.045 |
| CR383676.1 | 0.056 | id1 | -0.044 |
| cspg5a | 0.055 | cks1b | -0.044 |
| marcksl1b | 0.055 | ahcy | -0.044 |
| zgc:77784 | 0.054 | lig1 | -0.044 |
| cdh30 | 0.053 | snu13b | -0.044 |
| slitrk6 | 0.053 | rpa2 | -0.044 |
| mpp6b | 0.053 | zgc:56493 | -0.044 |
| tecrl2b | 0.052 | hmgb2a | -0.044 |
| fzd9a | 0.052 | msi1 | -0.043 |
| pcp4l1 | 0.052 | mcm6 | -0.043 |
| sox4a | 0.052 | nasp | -0.043 |
| vsx1 | 0.051 | COX7A2 (1 of many) | -0.043 |
| ppdpfb | 0.051 | cldn5a | -0.042 |
| myt1b | 0.051 | rrm2 | -0.042 |




