protocadherin 1 gamma 14
ZFIN 
Other cell groups


all cells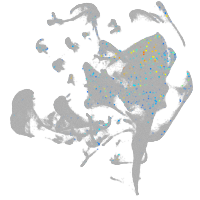 |
blastomeres |
PGCs |
endoderm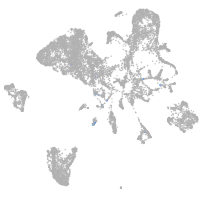 |
axial mesoderm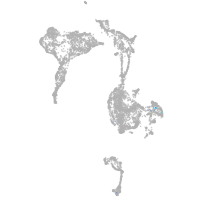 |
muscle 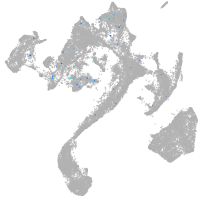 |
mural cells / non-skeletal muscle  |
mesenchyme 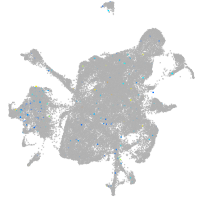 |
hematopoietic / vasculature 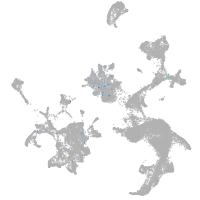 |
pronephros 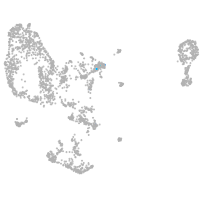 |
neural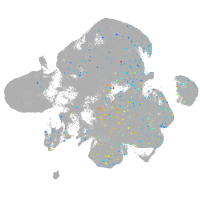 |
spinal cord / glia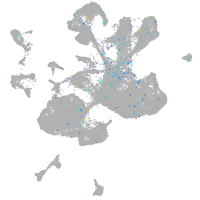 |
eye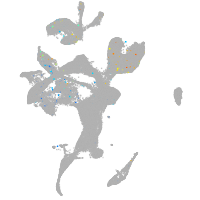 |
taste / olfactory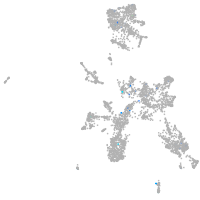 |
otic / lateral line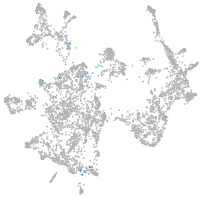 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-233f7.6 | 0.678 | sec61b | -0.044 |
| ankrd6a | 0.659 | rpl18a | -0.042 |
| LOC101882067 | 0.457 | tmed2 | -0.041 |
| crfb2 | 0.455 | rpl12 | -0.041 |
| prdm1b | 0.451 | rpl23 | -0.038 |
| wnt6a | 0.435 | hdlbpa | -0.038 |
| XLOC-014477 | 0.416 | rpl32 | -0.038 |
| irx6a | 0.401 | sec61g | -0.036 |
| veph1 | 0.394 | ssr4 | -0.036 |
| igf1 | 0.354 | kdelr2a | -0.036 |
| brinp3a.2 | 0.354 | rps18 | -0.035 |
| dmgdh | 0.323 | ube2v2 | -0.034 |
| dab1b | 0.309 | dad1 | -0.033 |
| stxbp1b | 0.304 | asph | -0.033 |
| hcst | 0.300 | ppp2cb | -0.032 |
| crmp1 | 0.292 | tpt1 | -0.032 |
| pcdh1gb2 | 0.292 | arf5 | -0.032 |
| FAM163A | 0.290 | tram1 | -0.031 |
| nectin1a | 0.275 | lrrc59 | -0.031 |
| zgc:153441 | 0.275 | dnajc1 | -0.030 |
| pitpnab | 0.273 | ccng1 | -0.030 |
| BX119963.1 | 0.269 | calr3b | -0.029 |
| crx | 0.252 | cfl1 | -0.029 |
| six6b | 0.248 | tmed10 | -0.028 |
| fezf1 | 0.241 | rps5 | -0.028 |
| rtn2a | 0.240 | hypk | -0.028 |
| ano8b | 0.239 | ssr2 | -0.028 |
| crtc1b | 0.237 | hspb1 | -0.028 |
| ttyh1 | 0.231 | pfn2l | -0.028 |
| CU234171.1 | 0.228 | rpl4 | -0.028 |
| zgc:158254 | 0.225 | rrbp1b | -0.027 |
| plekho1a | 0.224 | sept15 | -0.027 |
| tspoap1 | 0.218 | anp32b | -0.027 |
| pcdh11 | 0.216 | slc39a7 | -0.027 |
| trim3a | 0.216 | qkia | -0.027 |




