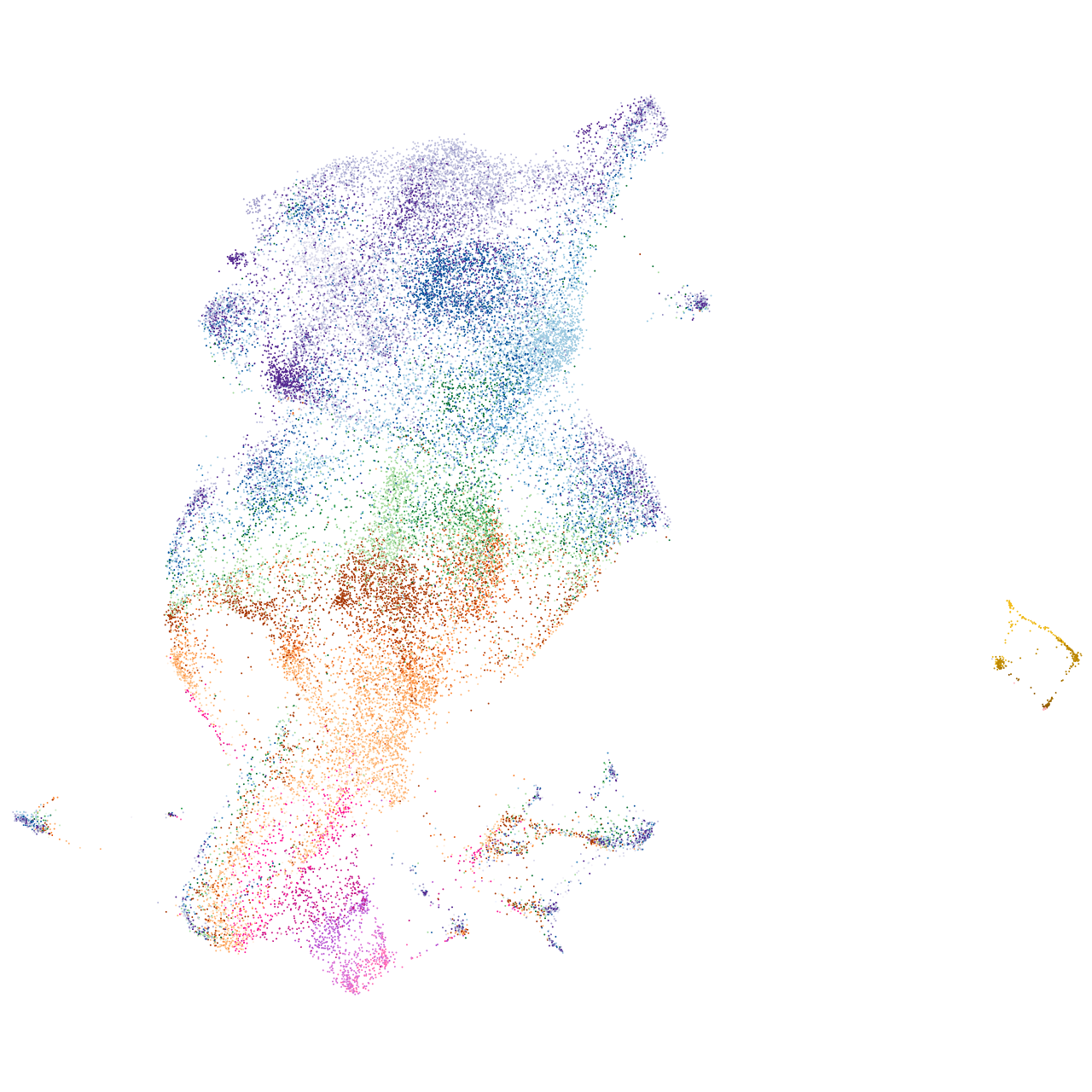pantothenate kinase 1b
ZFIN 
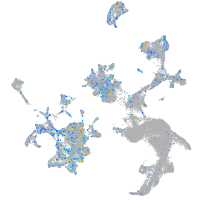
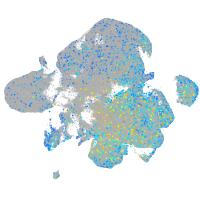
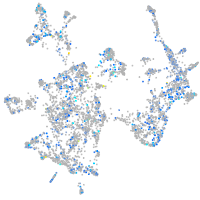
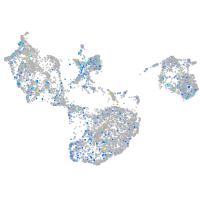
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR383676.1 | 0.095 | cyt1 | -0.062 |
| NC-002333.4 | 0.084 | krt4 | -0.061 |
| COX3 | 0.080 | krt17 | -0.053 |
| mt-cyb | 0.078 | cyt1l | -0.051 |
| mt-co2 | 0.077 | krt5 | -0.048 |
| mt-co1 | 0.076 | rpl7 | -0.041 |
| mt-nd1 | 0.075 | btf3 | -0.040 |
| golgb1 | 0.074 | rpl8 | -0.037 |
| LOC103909086 | 0.071 | rpl21 | -0.036 |
| mt-atp6 | 0.070 | rpl7a | -0.034 |
| pnrc2 | 0.066 | si:ch211-195b11.3 | -0.034 |
| nol4lb | 0.064 | faua | -0.034 |
| NC-002333.17 | 0.063 | ahcy | -0.033 |
| mt-nd2 | 0.062 | eef1a1l1 | -0.033 |
| clvs2 | 0.062 | rps2 | -0.032 |
| pleca | 0.061 | rplp0 | -0.032 |
| CR774179.5 | 0.061 | rpl10a | -0.032 |
| pbx4 | 0.061 | krtt1c19e | -0.031 |
| LOC108190033 | 0.061 | rplp1 | -0.031 |
| tent5ba | 0.061 | rpl35 | -0.030 |
| rab10 | 0.060 | rpl19 | -0.029 |
| ptmab | 0.060 | romo1 | -0.029 |
| pak6a | 0.060 | krt91 | -0.029 |
| zgc:111986 | 0.060 | rpl17 | -0.029 |
| smarca4a | 0.059 | actb2 | -0.028 |
| LO017711.1 | 0.059 | zgc:193505 | -0.028 |
| gipr | 0.059 | rps7 | -0.028 |
| LOC110437924.1 | 0.059 | rsl24d1 | -0.028 |
| tm9sf3 | 0.059 | gsta.1 | -0.028 |
| CABZ01060891.1 | 0.057 | rps24 | -0.027 |
| aco1 | 0.057 | rps26l | -0.026 |
| acin1a | 0.057 | s100a10b | -0.026 |
| tuba1c | 0.057 | anxa2a | -0.026 |
| calr3b | 0.057 | rps23 | -0.025 |
| hsp90b1 | 0.056 | eevs | -0.025 |