pantothenate kinase 1b
ZFIN 
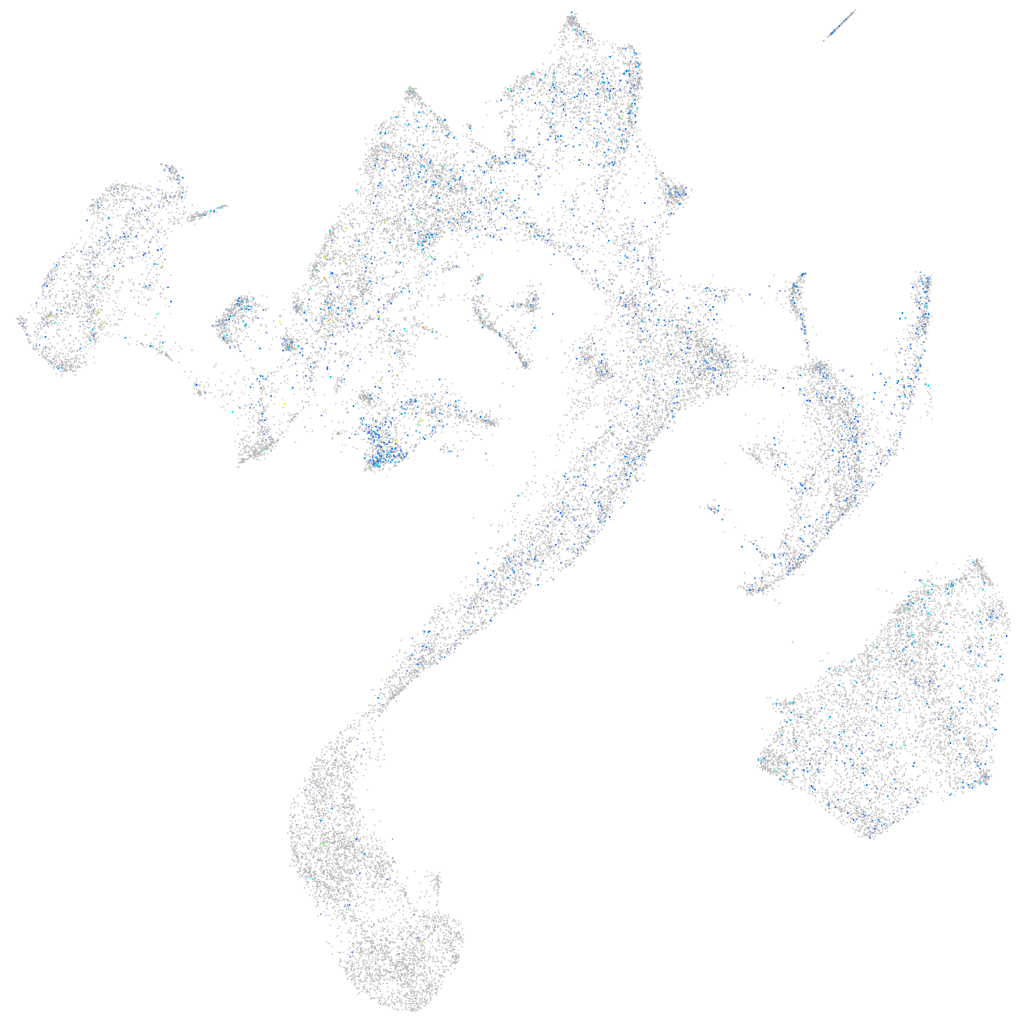
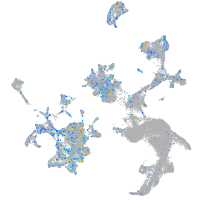
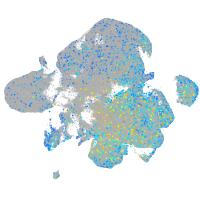
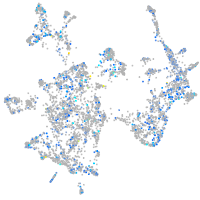
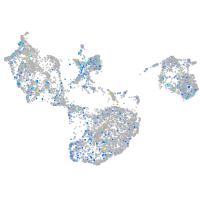
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tubb5 | 0.106 | myoz1b | -0.071 |
| gng3 | 0.100 | slc25a4 | -0.070 |
| gpm6aa | 0.095 | myom2a | -0.069 |
| rtn1b | 0.089 | mylpfb | -0.069 |
| atp6v0cb | 0.088 | si:ch211-255p10.3 | -0.067 |
| tuba1c | 0.087 | casq1a | -0.067 |
| stmn1b | 0.087 | si:ch73-367p23.2 | -0.067 |
| vamp2 | 0.086 | eef2l2 | -0.067 |
| stxbp1a | 0.086 | ryr3 | -0.064 |
| kif1ab | 0.085 | CABZ01061524.1 | -0.064 |
| ptmaa | 0.084 | hhatla | -0.063 |
| elavl3 | 0.084 | actn3a | -0.063 |
| sncb | 0.084 | tnni2a.4 | -0.063 |
| ckbb | 0.082 | tnnt3b | -0.063 |
| gap43 | 0.082 | mylz3 | -0.062 |
| gabrb4 | 0.082 | atp2a1l | -0.061 |
| elavl4 | 0.081 | XLOC-005350 | -0.061 |
| nova2 | 0.080 | ampd1 | -0.061 |
| cspg5a | 0.080 | tmod4 | -0.061 |
| cdk5r1b | 0.078 | XLOC-025819 | -0.061 |
| fez1 | 0.078 | cdnf | -0.060 |
| gnao1a | 0.078 | XLOC-001975 | -0.060 |
| myt1b | 0.077 | myl1 | -0.060 |
| atp1a3a | 0.077 | pvalb1 | -0.060 |
| evlb | 0.077 | XLOC-006515 | -0.060 |
| cfl1 | 0.077 | smyd1a | -0.060 |
| atpv0e2 | 0.077 | pvalb2 | -0.059 |
| hspa5 | 0.077 | hsc70 | -0.059 |
| stmn2a | 0.076 | myom1a | -0.058 |
| gapdhs | 0.076 | myhz1.1 | -0.057 |
| jpt1b | 0.075 | pkmb | -0.057 |
| calr | 0.075 | prr33 | -0.057 |
| atp6v1g1 | 0.074 | myhz1.3 | -0.056 |
| actb2 | 0.073 | dhrs7cb | -0.056 |
| stx1b | 0.073 | myoz2a | -0.056 |