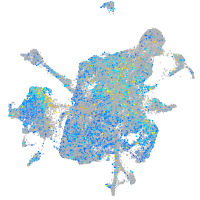
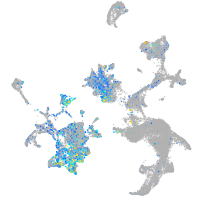
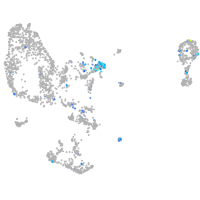
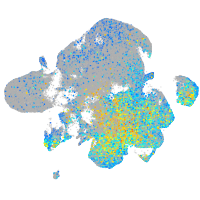
paralemmin 1b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.545 | si:dkey-151g10.6 | -0.186 |
| sncb | 0.504 | atp1b1a | -0.182 |
| stmn1b | 0.470 | cdh17 | -0.174 |
| stmn2a | 0.468 | cox7b | -0.168 |
| ywhag2 | 0.466 | rbm47 | -0.163 |
| csdc2a | 0.463 | ezra | -0.160 |
| dnajc5aa | 0.459 | sgk1 | -0.158 |
| mllt11 | 0.452 | suclg1 | -0.157 |
| rtn1a | 0.445 | COX5B | -0.156 |
| stxbp1a | 0.444 | uqcrq | -0.154 |
| zgc:65894 | 0.438 | atp1a1a.4 | -0.153 |
| gap43 | 0.438 | aldh6a1 | -0.151 |
| atp6v0cb | 0.434 | sypl2a | -0.150 |
| stx1b | 0.429 | COX7A2 (1 of many) | -0.148 |
| rtn1b | 0.426 | zgc:165573 | -0.146 |
| gpm6ab | 0.418 | slc25a5 | -0.145 |
| snap25a | 0.417 | acy3.2 | -0.144 |
| zgc:153426 | 0.416 | uqcrh | -0.143 |
| si:dkeyp-75h12.5 | 0.414 | clic5b | -0.143 |
| vamp2 | 0.414 | hnf1ba | -0.141 |
| tubb5 | 0.408 | eno3 | -0.140 |
| si:dkey-276j7.1 | 0.404 | tmprss2 | -0.140 |
| cnrip1a | 0.403 | atp5pf | -0.137 |
| tuba1c | 0.401 | ehd1b | -0.137 |
| celf2 | 0.398 | cox7c | -0.137 |
| id4 | 0.398 | atp5fa1 | -0.137 |
| gpm6aa | 0.398 | rps28 | -0.136 |
| eno2 | 0.395 | atp5f1c | -0.136 |
| chd5 | 0.392 | mrpl36 | -0.136 |
| nsg2 | 0.392 | chp1 | -0.136 |
| elavl4 | 0.391 | rps2 | -0.135 |
| scg2b | 0.390 | rpl35a | -0.135 |
| fez1 | 0.388 | rpl12 | -0.135 |
| sv2a | 0.387 | rnaseka | -0.134 |
| elavl3 | 0.386 | degs1 | -0.132 |