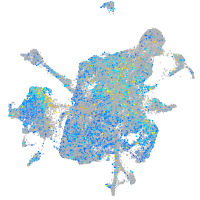
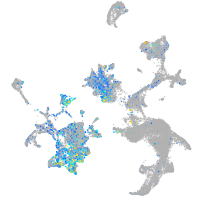
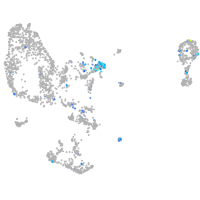
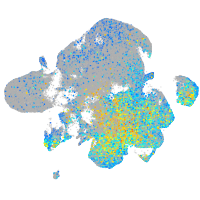
paralemmin 1b
ZFIN 
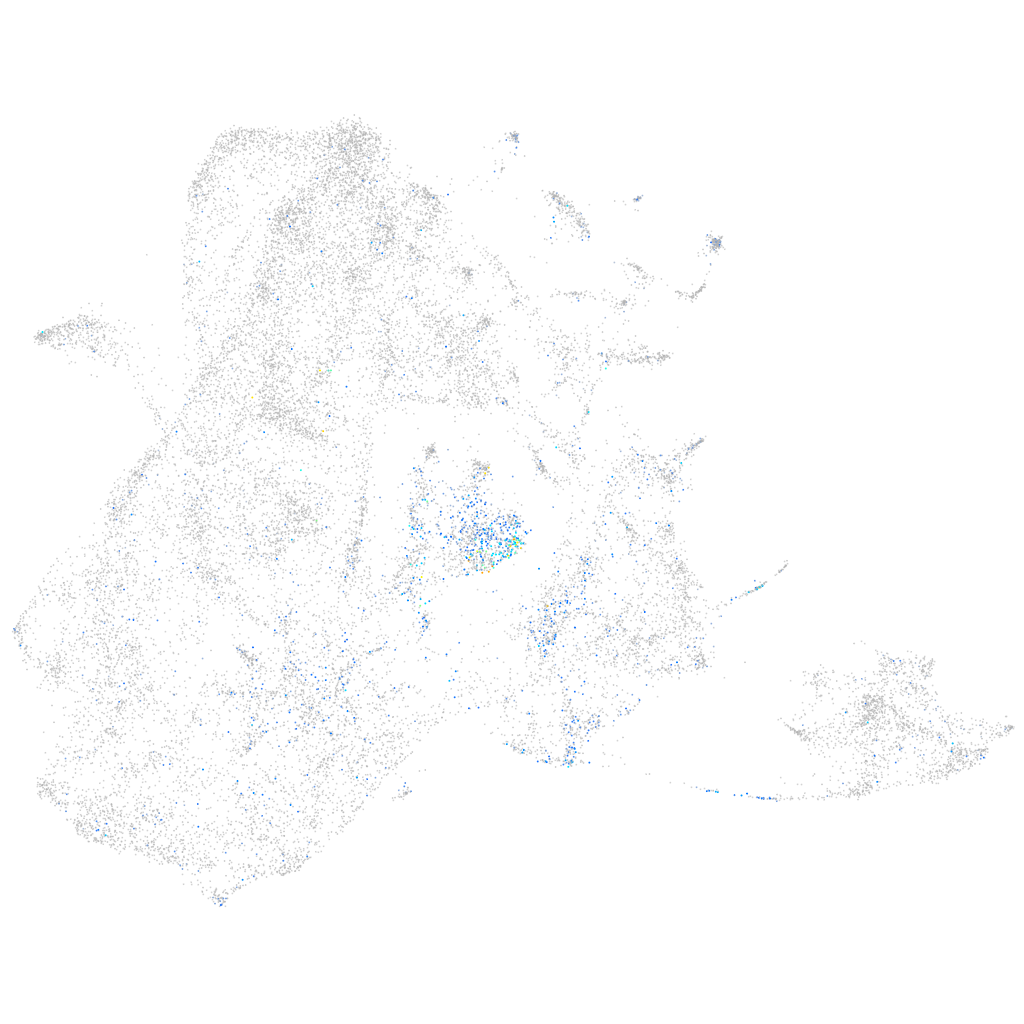
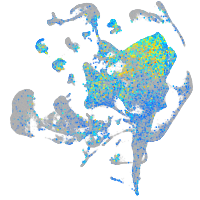
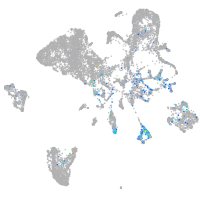
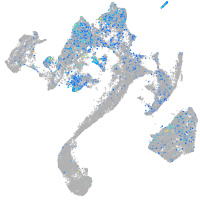
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.303 | cfl1l | -0.168 |
| sncb | 0.302 | tmsb1 | -0.155 |
| snap25a | 0.295 | pfn1 | -0.154 |
| atp6v0cb | 0.287 | krt4 | -0.146 |
| gpm6ab | 0.287 | cyt1 | -0.136 |
| stmn1b | 0.281 | s100a10b | -0.122 |
| ywhag2 | 0.279 | cldni | -0.111 |
| stxbp1a | 0.278 | actb1 | -0.111 |
| gpm6aa | 0.273 | spaca4l | -0.110 |
| rtn1a | 0.272 | cldn1 | -0.107 |
| elavl4 | 0.270 | actb2 | -0.102 |
| tuba1c | 0.260 | cyt1l | -0.102 |
| map1aa | 0.259 | capns1a | -0.101 |
| rtn1b | 0.259 | col11a1a | -0.098 |
| elavl3 | 0.256 | anxa1a | -0.097 |
| stmn2a | 0.255 | ecrg4b | -0.097 |
| vamp2 | 0.255 | epcam | -0.095 |
| olfm1b | 0.254 | lgals3b | -0.094 |
| gnao1a | 0.254 | ahnak | -0.094 |
| nova2 | 0.251 | zgc:162730 | -0.092 |
| stx1b | 0.248 | si:dkey-16p21.8 | -0.092 |
| eno2 | 0.247 | zgc:175088 | -0.092 |
| sncgb | 0.246 | hdlbpa | -0.091 |
| cspg5a | 0.245 | col1a2 | -0.091 |
| zgc:65894 | 0.240 | cap1 | -0.091 |
| zgc:153426 | 0.239 | tuba8l2 | -0.091 |
| tuba2 | 0.238 | cebpd | -0.089 |
| tmsb2 | 0.237 | col2a1b | -0.089 |
| fez1 | 0.232 | thy1 | -0.089 |
| syn2a | 0.232 | col18a1a | -0.088 |
| atpv0e2 | 0.229 | krt5 | -0.088 |
| syt1a | 0.229 | pleca | -0.088 |
| mllt11 | 0.229 | wu:fb18f06 | -0.087 |
| st8sia5 | 0.228 | sparc | -0.086 |
| tmem59l | 0.227 | col1a1b | -0.086 |