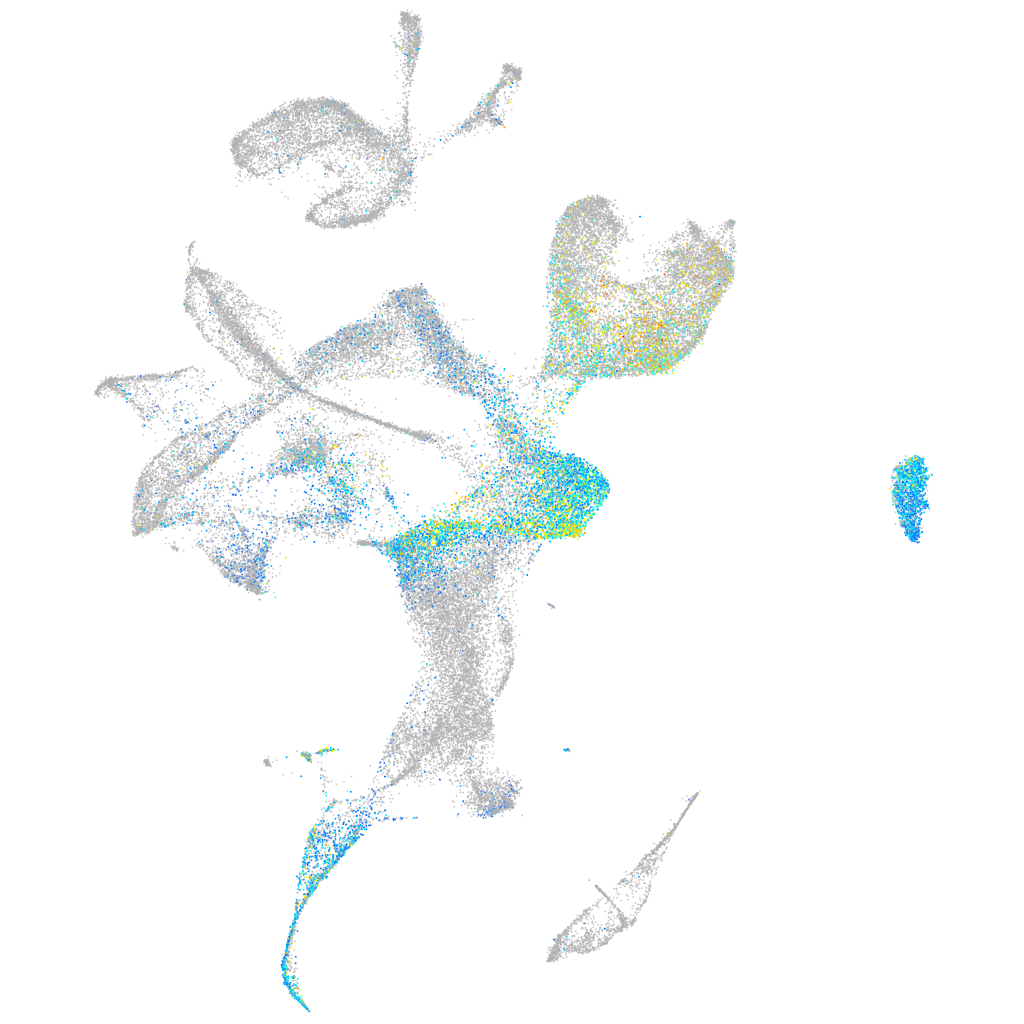
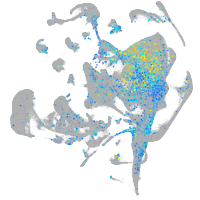
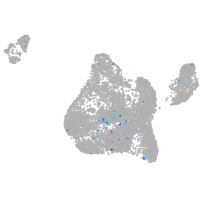
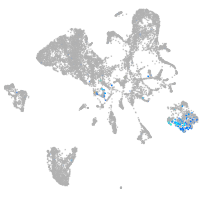
orthodenticle homeobox 2b
ZFIN 
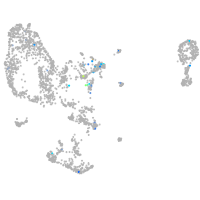
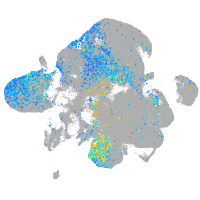
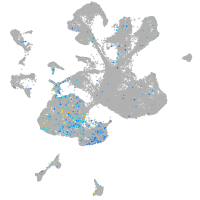
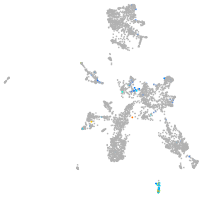
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| olig2 | 0.366 | gapdhs | -0.220 |
| im:7152348 | 0.319 | sncb | -0.164 |
| insm1a | 0.298 | atp6v0cb | -0.156 |
| neurog1 | 0.294 | mt-atp6 | -0.155 |
| hes2.1 | 0.282 | atp5mc1 | -0.152 |
| foxn4 | 0.269 | pax6b | -0.152 |
| tp53inp2 | 0.265 | snap25b | -0.144 |
| hes2.2 | 0.257 | elavl3 | -0.144 |
| ptprua | 0.255 | mt-nd1 | -0.143 |
| neurod4 | 0.251 | cox7a2a | -0.137 |
| hes6 | 0.250 | vamp2 | -0.136 |
| sox3 | 0.247 | atp5if1b | -0.135 |
| pif1 | 0.246 | spock3 | -0.134 |
| birc5a | 0.241 | pax6a | -0.133 |
| CU634008.1 | 0.241 | c1qbp | -0.130 |
| nusap1 | 0.239 | eno1a | -0.130 |
| gadd45ga | 0.237 | chchd10 | -0.130 |
| ube2c | 0.233 | vdac1 | -0.128 |
| plk1 | 0.231 | atpv0e2 | -0.126 |
| hmgb2a | 0.228 | sypb | -0.126 |
| msna | 0.226 | mab21l1 | -0.124 |
| tpx2 | 0.225 | gng3 | -0.124 |
| si:ch211-69g19.2 | 0.222 | atp5pf | -0.123 |
| mki67 | 0.221 | elovl4b | -0.121 |
| aspm | 0.217 | atp5mc3b | -0.121 |
| sept4a | 0.216 | si:dkey-16p21.8 | -0.120 |
| kif11 | 0.215 | hint1 | -0.119 |
| prc1a | 0.214 | cox8a | -0.118 |
| cenpf | 0.213 | tpi1b | -0.118 |
| dlgap5 | 0.211 | stxbp1a | -0.117 |
| kifc1 | 0.211 | syt1a | -0.117 |
| lbr | 0.209 | ldhba | -0.116 |
| tuba1b | 0.209 | sh3gl2a | -0.115 |
| vsx1 | 0.208 | cox6a1 | -0.114 |
| kif23 | 0.207 | rx2 | -0.114 |