orthodenticle homeobox 1
ZFIN 
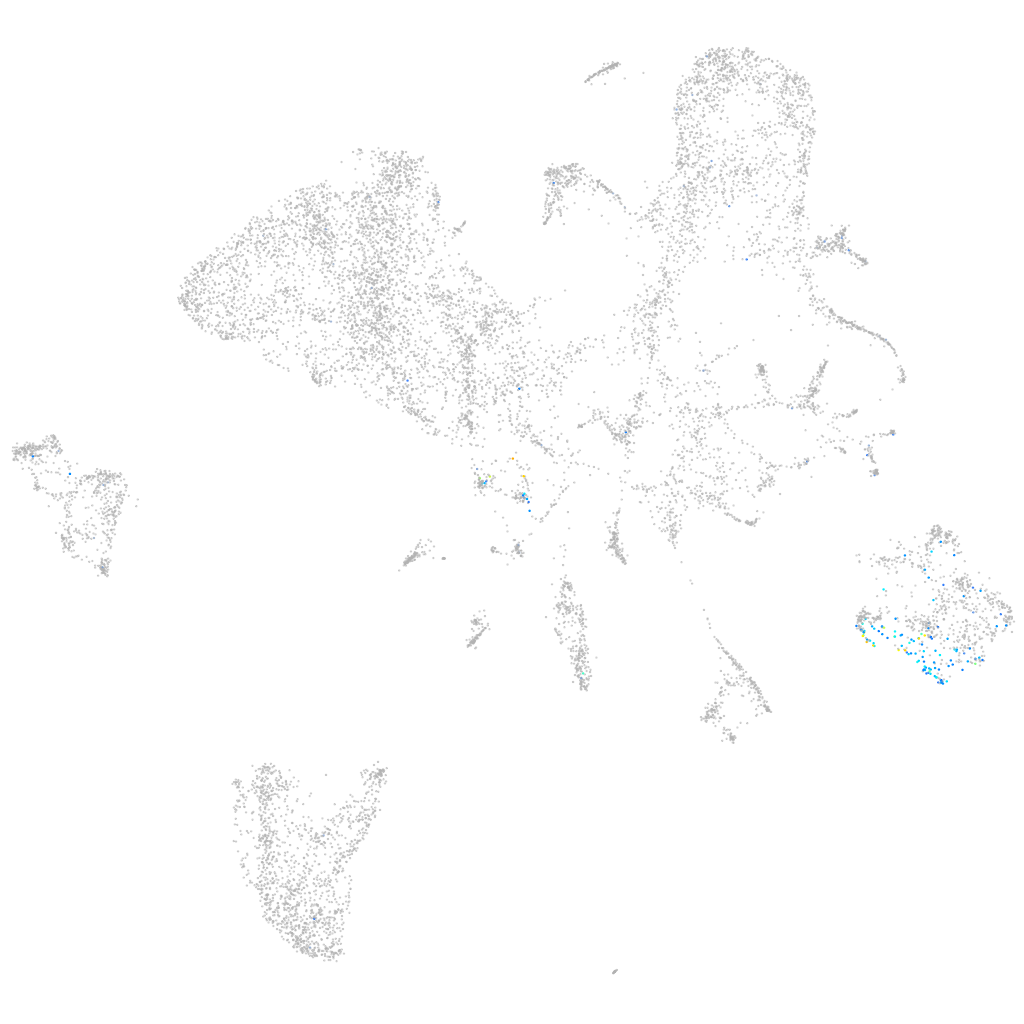
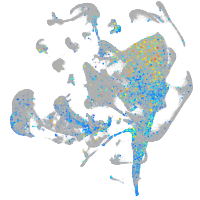
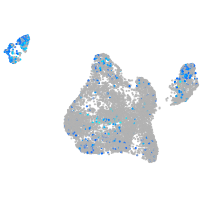
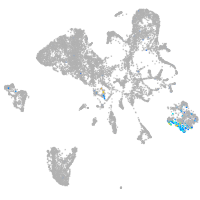
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gsc | 0.506 | rps10 | -0.253 |
| dmbx1a | 0.461 | rpl37 | -0.249 |
| shisa2a | 0.421 | zgc:114188 | -0.236 |
| cyp26c1 | 0.409 | rps17 | -0.213 |
| otx2a | 0.395 | nme2b.1 | -0.169 |
| otx2b | 0.376 | zgc:56493 | -0.153 |
| nog1 | 0.374 | zgc:92744 | -0.137 |
| fzd8b | 0.355 | eno3 | -0.135 |
| her5 | 0.344 | prdx2 | -0.134 |
| lft2 | 0.330 | ahcy | -0.132 |
| si:dkey-43p13.5 | 0.304 | atp5f1b | -0.131 |
| si:ch211-152c2.3 | 0.302 | atp5mc3b | -0.130 |
| akap12b | 0.294 | mt-nd1 | -0.129 |
| fscn1a | 0.288 | rpl9 | -0.126 |
| marcksl1b | 0.281 | atp5l | -0.126 |
| lft1 | 0.281 | eef1da | -0.126 |
| iqca1 | 0.281 | sod1 | -0.126 |
| sfrp1a | 0.272 | suclg1 | -0.125 |
| pkdcca | 0.258 | atp5if1b | -0.125 |
| irx7 | 0.257 | cox6a1 | -0.125 |
| cxcr4a | 0.256 | COX7A2 (1 of many) | -0.123 |
| ndr2 | 0.256 | zgc:158463 | -0.122 |
| asph | 0.255 | atp5fa1 | -0.122 |
| nes | 0.252 | atp5pd | -0.121 |
| marcksb | 0.251 | romo1 | -0.119 |
| ppig | 0.249 | gapdh | -0.118 |
| sox32 | 0.244 | gamt | -0.118 |
| BX649529.1 | 0.242 | ndufb10 | -0.118 |
| flrt3 | 0.242 | gstp1 | -0.117 |
| tbx1 | 0.238 | eef1b2 | -0.116 |
| noto | 0.237 | atp5meb | -0.115 |
| cxcl12b | 0.236 | rplp0 | -0.114 |
| NC-002333.4 | 0.235 | atp5mc1 | -0.113 |
| hnrnpa1b | 0.235 | atp5pf | -0.112 |
| snrnp70 | 0.234 | rps20 | -0.111 |