otogelin
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
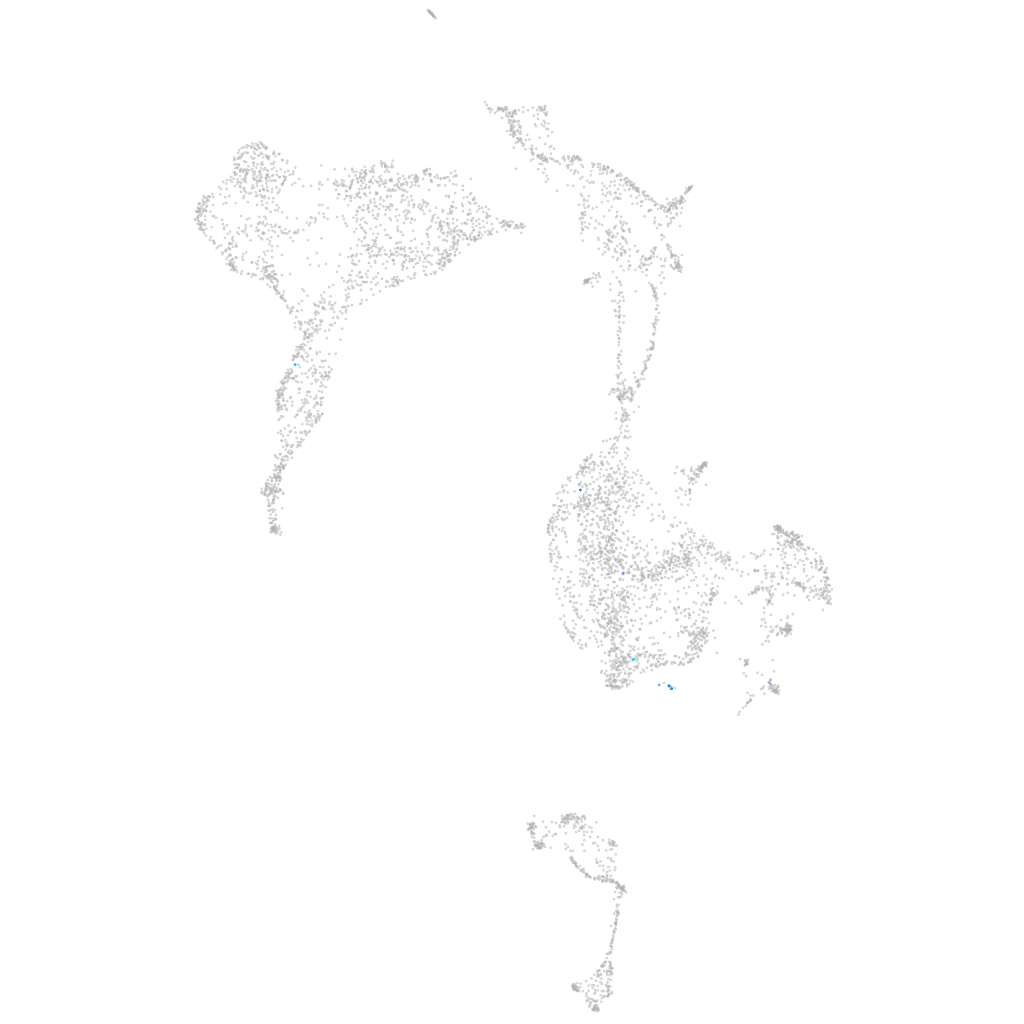
endoderm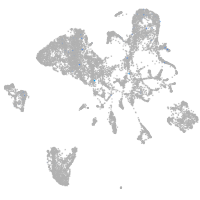 |
axial mesoderm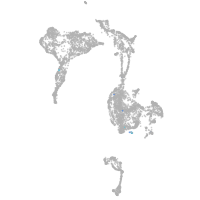 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 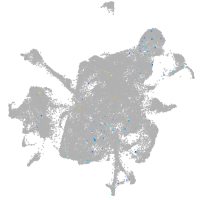 |
hematopoietic / vasculature 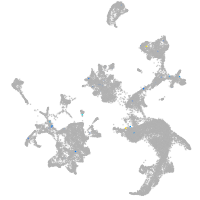 |
pronephros 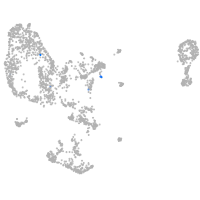 |
neural |
spinal cord / glia |
eye |
taste / olfactory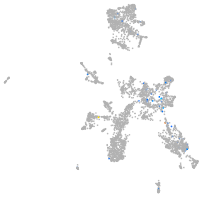 |
otic / lateral line |
epidermis |
periderm |
fin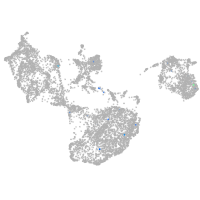 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zpld1a | 0.657 | zgc:193541 | -0.039 |
| zpld1b | 0.642 | psmb3 | -0.037 |
| LOC100331291 | 0.495 | rap1aa | -0.034 |
| cryabb | 0.475 | snrpd1 | -0.034 |
| slc25a22b | 0.469 | psmb7 | -0.032 |
| CR383669.1 | 0.422 | erh | -0.032 |
| XLOC-034214 | 0.422 | hmga1a | -0.031 |
| leg1.2 | 0.415 | zgc:86598 | -0.031 |
| si:dkey-1m11.6 | 0.414 | cox7a2a | -0.030 |
| si:dkey-222f2.1 | 0.412 | ppp2cb | -0.029 |
| mb | 0.403 | sept6 | -0.029 |
| hhla1 | 0.378 | si:dkey-56m19.5 | -0.028 |
| slc13a5a | 0.371 | hnrnpa1a | -0.028 |
| capn3a | 0.357 | dnajc1 | -0.028 |
| ifit10 | 0.350 | tcea1 | -0.028 |
| atp1a1a.4 | 0.323 | hmgn6 | -0.027 |
| CR388148.2 | 0.319 | h2afva | -0.027 |
| ndnfl | 0.318 | hmgn2 | -0.027 |
| vwa10.1 | 0.301 | tubb2b | -0.027 |
| rln3a | 0.290 | ppm1g | -0.027 |
| kcnq5a | 0.284 | nip7 | -0.027 |
| CU459155.1 | 0.277 | chaf1a | -0.027 |
| ms4a17a.2 | 0.264 | pinx1 | -0.027 |
| slc16a1a | 0.258 | tram1 | -0.027 |
| tnni3k | 0.255 | ube2d2 | -0.027 |
| si:dkey-27j5.5 | 0.253 | fkbp11 | -0.026 |
| zgc:112437 | 0.251 | gspt1l | -0.026 |
| six1b | 0.248 | actl6a | -0.026 |
| zwi | 0.238 | bms1 | -0.026 |
| tmprss7 | 0.238 | gtf2f1 | -0.026 |
| zgc:194312 | 0.227 | psma6a | -0.026 |
| slc26a4 | 0.227 | ssb | -0.026 |
| slc7a8b | 0.226 | chchd2 | -0.026 |
| si:ch211-236l14.4 | 0.222 | hmgb2a | -0.026 |
| si:dkey-98f17.3 | 0.214 | zgc:110216 | -0.026 |