O-GlcNAcase
ZFIN 
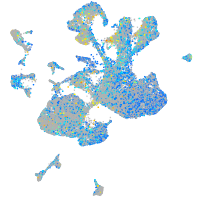
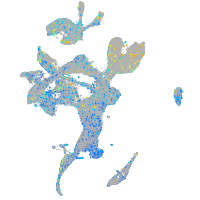
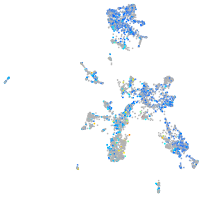
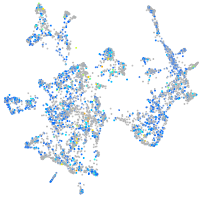
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.093 | fabp7a | -0.080 |
| CABZ01075068.1 | 0.086 | atp1a1b | -0.063 |
| myt1a | 0.080 | pvalb1 | -0.062 |
| trim71 | 0.077 | CU467822.1 | -0.058 |
| nova2 | 0.075 | ppap2d | -0.055 |
| dlb | 0.072 | pvalb2 | -0.054 |
| slc16a9b | 0.070 | slc1a2b | -0.054 |
| tp53inp2 | 0.068 | actc1b | -0.053 |
| hoxb3a | 0.068 | efhd1 | -0.053 |
| hoxc3a | 0.067 | glula | -0.051 |
| NC-002333.4 | 0.066 | atp1b4 | -0.050 |
| notch1a | 0.065 | si:ch211-156b7.4 | -0.049 |
| gse1 | 0.065 | cdo1 | -0.048 |
| baz2ba | 0.063 | CR848047.1 | -0.046 |
| myt1b | 0.063 | sod1 | -0.044 |
| pnrc2 | 0.062 | ppa1b | -0.043 |
| ebf2 | 0.062 | acbd7 | -0.043 |
| lin28a | 0.061 | apoa1b | -0.042 |
| nr6a1a | 0.061 | mt2 | -0.042 |
| greb1l | 0.061 | apoa2 | -0.041 |
| cep131 | 0.061 | cd82a | -0.041 |
| hoxb1b | 0.061 | si:ch211-66e2.5 | -0.040 |
| mxi1 | 0.060 | mdkb | -0.040 |
| pbx4 | 0.060 | prss59.2 | -0.040 |
| igf2bp1 | 0.060 | cebpd | -0.039 |
| kdm5bb | 0.059 | nog2 | -0.039 |
| srrm4 | 0.059 | syngr2b | -0.039 |
| si:ch73-386h18.1 | 0.059 | elovl2 | -0.038 |
| nhlh2 | 0.058 | ckbb | -0.038 |
| neurod4 | 0.058 | zgc:165461 | -0.038 |
| chd7 | 0.058 | ptgdsb.2 | -0.038 |
| fstl1a | 0.058 | ddt | -0.038 |
| hsp90ab1 | 0.058 | crabp1a | -0.038 |
| si:ch211-222l21.1 | 0.057 | mylpfa | -0.038 |
| zswim5 | 0.057 | zgc:162944 | -0.037 |