nucleoporin 214
ZFIN 
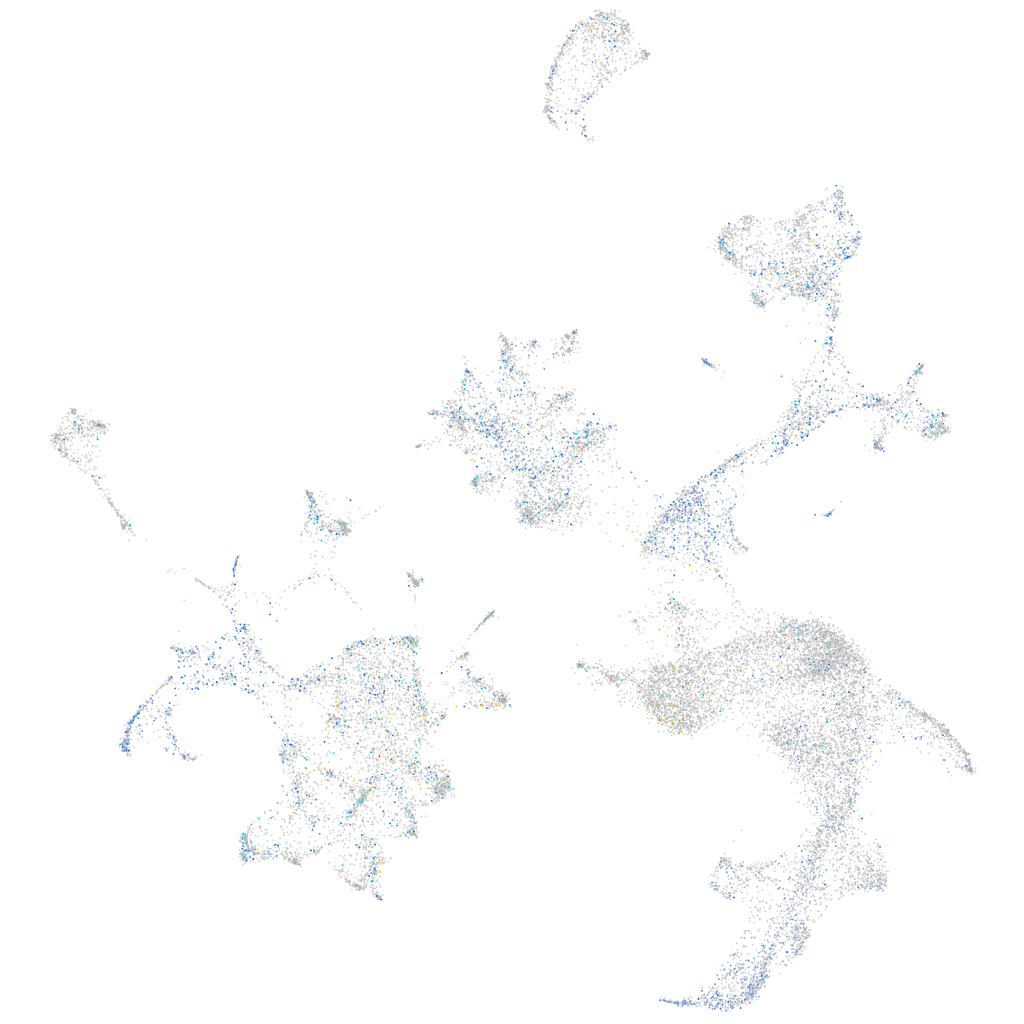
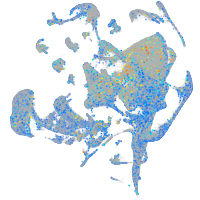
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mt-atp6 | 0.095 | hbbe2 | -0.068 |
| mt-nd1 | 0.094 | cahz | -0.066 |
| mt-nd2 | 0.093 | hbae3 | -0.066 |
| mt-co2 | 0.092 | plac8l1 | -0.064 |
| hmgb2b | 0.092 | hbbe1.3 | -0.064 |
| mt-co1 | 0.091 | hbae1.3 | -0.064 |
| mt-cyb | 0.091 | hbbe1.1 | -0.062 |
| snrpf | 0.090 | hbbe1.2 | -0.061 |
| h3f3a | 0.090 | hemgn | -0.060 |
| hsp90ab1 | 0.089 | hbae1.1 | -0.059 |
| tubb2b | 0.089 | gpx1a | -0.059 |
| khdrbs1a | 0.089 | nt5c2l1 | -0.058 |
| rpl6 | 0.089 | tmod4 | -0.058 |
| snrpg | 0.088 | creg1 | -0.058 |
| hmga1a | 0.087 | si:ch211-250g4.3 | -0.056 |
| tuba8l | 0.087 | tspo | -0.056 |
| h2afvb | 0.086 | blvrb | -0.052 |
| ranbp1 | 0.086 | selenoj | -0.051 |
| hnrnpa0l | 0.086 | alas2 | -0.051 |
| rbm8a | 0.085 | si:ch211-207c6.2 | -0.047 |
| sub1a | 0.085 | zgc:163057 | -0.044 |
| COX3 | 0.085 | prdx2 | -0.044 |
| psma3 | 0.084 | fth1a | -0.043 |
| atp5fa1 | 0.084 | hif1al2 | -0.040 |
| snrpd1 | 0.084 | si:dkey-240n22.3 | -0.039 |
| rpl36a | 0.084 | nprl3 | -0.038 |
| mt-nd4 | 0.084 | gabarapa | -0.037 |
| rplp2l | 0.084 | gp9 | -0.036 |
| rpl23 | 0.084 | bloc1s6 | -0.035 |
| rps7 | 0.084 | hbae5 | -0.035 |
| syncrip | 0.084 | eef2l2 | -0.034 |
| nutf2l | 0.083 | ucp3 | -0.034 |
| rps27.1 | 0.083 | zgc:56095 | -0.033 |
| snu13b | 0.083 | fbxo32 | -0.032 |
| rpl13 | 0.083 | si:ch211-103n10.5 | -0.031 |