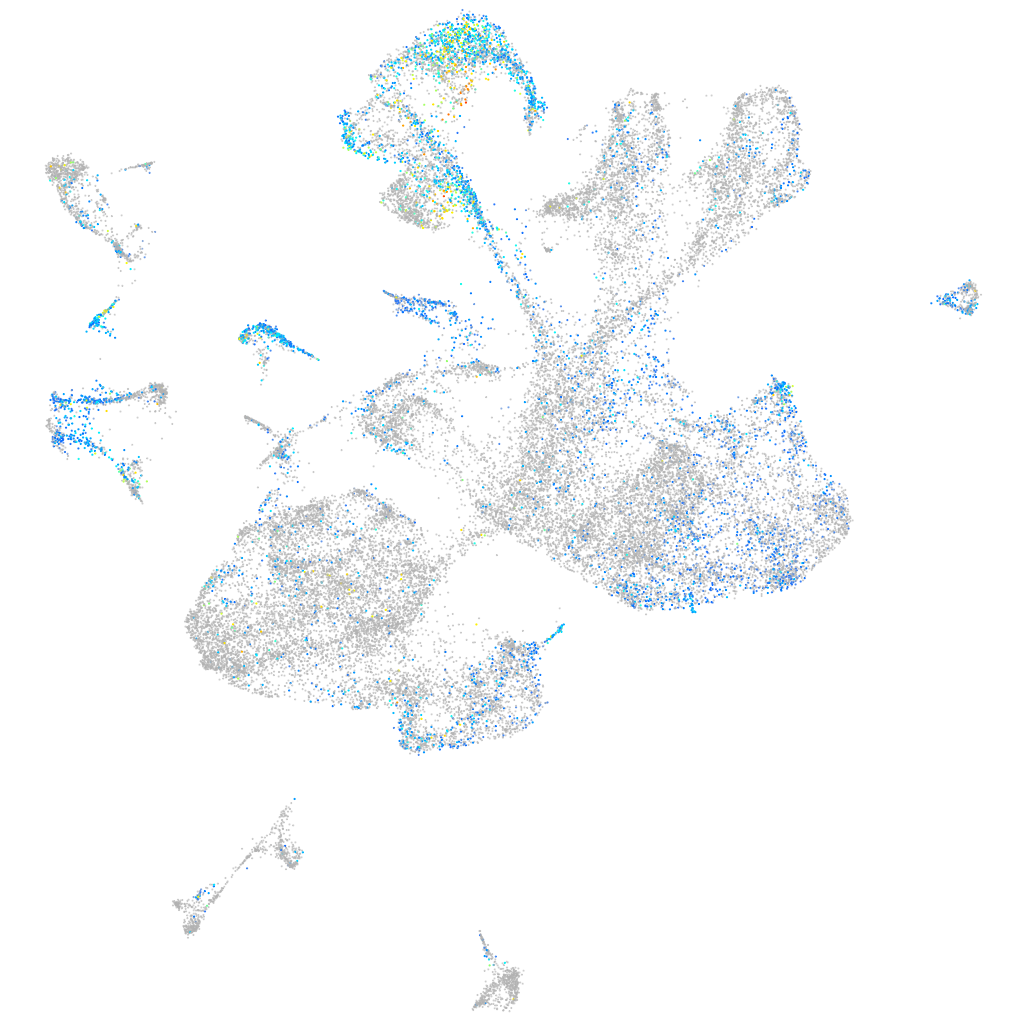
neuropilin 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slit3 | 0.338 | CU467822.1 | -0.172 |
| chodl | 0.300 | fabp7a | -0.154 |
| isl2a | 0.255 | zgc:165461 | -0.151 |
| slc18a3a | 0.250 | sox3 | -0.146 |
| phox2a | 0.238 | CR848047.1 | -0.138 |
| phox2bb | 0.230 | gpm6bb | -0.136 |
| isl1 | 0.227 | atp1a1b | -0.134 |
| prph | 0.224 | CU634008.1 | -0.133 |
| onecut1 | 0.215 | slc1a2b | -0.133 |
| tbx20 | 0.210 | si:ch73-21g5.7 | -0.125 |
| islr2 | 0.196 | notch3 | -0.123 |
| bcam | 0.194 | XLOC-003690 | -0.123 |
| nefmb | 0.187 | atp1b4 | -0.122 |
| CR812832.1 | 0.179 | sox19a | -0.122 |
| agrn | 0.178 | cd82a | -0.120 |
| gap43 | 0.178 | si:ch211-251b21.1 | -0.118 |
| lmo4b | 0.178 | mdka | -0.114 |
| mnx1 | 0.173 | sb:cb81 | -0.112 |
| tbx2b | 0.171 | her4.2 | -0.110 |
| gab2 | 0.168 | plp1a | -0.106 |
| arid3c | 0.166 | her15.1 | -0.106 |
| shc2 | 0.163 | lfng | -0.105 |
| mmp17a | 0.163 | si:ch73-30b17.2 | -0.105 |
| ptprfa | 0.163 | XLOC-003689 | -0.105 |
| si:ch211-247j9.1 | 0.163 | id1 | -0.103 |
| map1b | 0.161 | nrarpb | -0.102 |
| slc5a7a | 0.161 | slc6a9 | -0.102 |
| hoxb6b | 0.158 | notch1b | -0.102 |
| hoxb5b | 0.157 | gas1b | -0.101 |
| sv2c | 0.154 | her4.1 | -0.098 |
| optc | 0.154 | msi1 | -0.098 |
| chata | 0.154 | her8a | -0.097 |
| lhx4 | 0.153 | ptprz1b | -0.093 |
| adcyap1b | 0.153 | s1pr1 | -0.092 |
| xpr1a | 0.151 | robo4 | -0.091 |