neuropilin 1a
ZFIN 
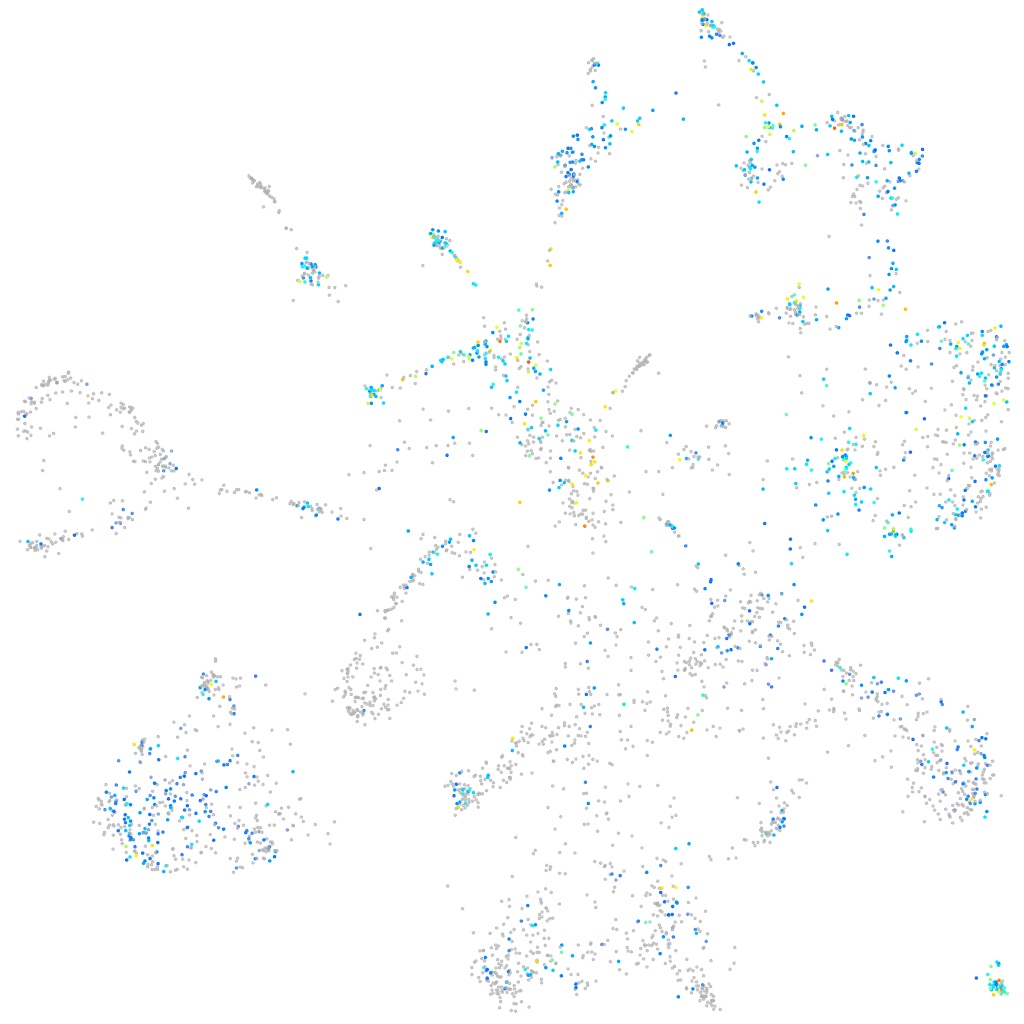
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt94 | 0.233 | si:ch211-62a1.3 | -0.205 |
| podxl | 0.216 | nkx2.3 | -0.197 |
| tmem88b | 0.215 | mylkb | -0.184 |
| efnb2a | 0.213 | col6a2 | -0.181 |
| gata5 | 0.206 | foxf1 | -0.172 |
| CR383676.1 | 0.204 | csrp1b | -0.167 |
| bnc2 | 0.197 | col6a1 | -0.165 |
| krt8 | 0.191 | gapdhs | -0.151 |
| akap12b | 0.190 | col4a6 | -0.151 |
| rbpms2b | 0.190 | cald1b | -0.150 |
| cdh11 | 0.189 | gpia | -0.149 |
| lurap1 | 0.189 | acta2 | -0.148 |
| jam2b | 0.185 | col4a5 | -0.146 |
| c1qtnf6a | 0.183 | ak1 | -0.146 |
| cxadr | 0.182 | myl6 | -0.145 |
| nid1a | 0.181 | angptl5 | -0.141 |
| cxcl12a | 0.179 | ntn5 | -0.141 |
| tenm3 | 0.178 | rprmb | -0.140 |
| epha7 | 0.176 | desmb | -0.140 |
| mmel1 | 0.176 | ckbb | -0.137 |
| wt1a | 0.175 | tagln | -0.135 |
| phactr4a | 0.175 | foxf2a | -0.134 |
| cdh2 | 0.174 | ldha | -0.134 |
| rai14 | 0.173 | rplp1 | -0.133 |
| gata6 | 0.172 | eno3 | -0.131 |
| cd151 | 0.171 | aldocb | -0.129 |
| jag1b | 0.169 | tpm2 | -0.128 |
| foxc1a | 0.168 | tpm1 | -0.125 |
| cyp1c1 | 0.168 | pgam1a | -0.124 |
| smad6a | 0.168 | tpi1b | -0.124 |
| arl4aa | 0.167 | cnn1b | -0.123 |
| slit3 | 0.166 | BX088707.3 | -0.121 |
| c7a | 0.166 | mgll | -0.121 |
| tbx18 | 0.165 | inhbaa | -0.120 |
| ism1 | 0.164 | eno1a | -0.120 |