"nuclear receptor subfamily 2, group F, member 5"
ZFIN 
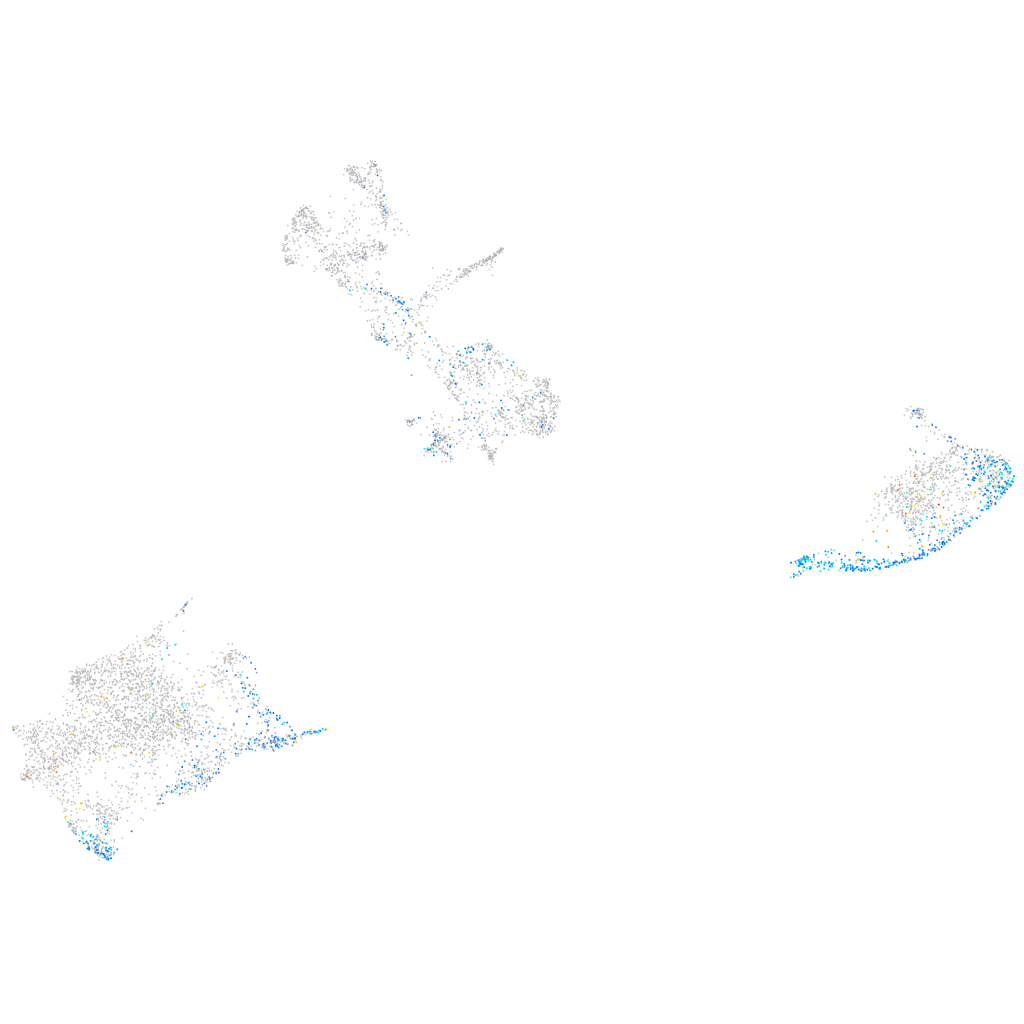
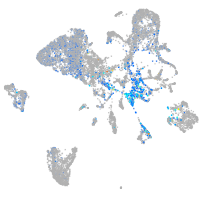
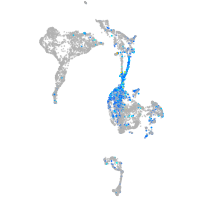
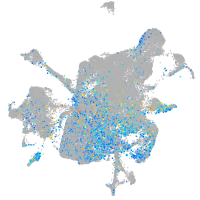
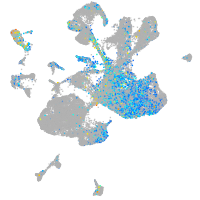
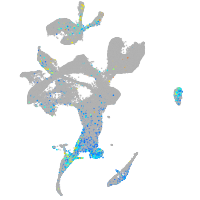
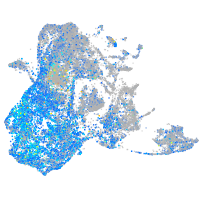
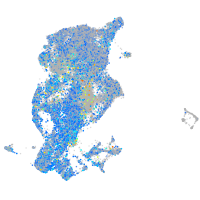
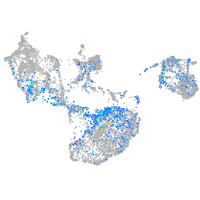
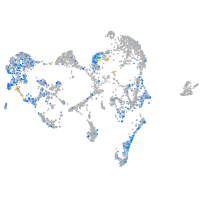
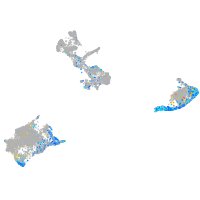
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msx1b | 0.309 | uraha | -0.175 |
| mitfa | 0.284 | paics | -0.142 |
| tmem243b | 0.279 | defbl1 | -0.138 |
| SPAG9 | 0.277 | apoda.1 | -0.131 |
| tfap2e | 0.275 | si:ch211-256m1.8 | -0.129 |
| tfap2a | 0.272 | si:dkey-251i10.2 | -0.129 |
| ednrab | 0.263 | gpnmb | -0.128 |
| lamp1a | 0.256 | lypc | -0.128 |
| crestin | 0.251 | CABZ01021592.1 | -0.126 |
| kita | 0.251 | mdh1aa | -0.126 |
| tyr | 0.243 | pvalb1 | -0.123 |
| slc1a3a | 0.241 | pvalb2 | -0.122 |
| rabl6b | 0.240 | hbbe1.3 | -0.116 |
| slc24a5 | 0.240 | phyhd1 | -0.116 |
| slc37a2 | 0.231 | si:dkey-197i20.6 | -0.114 |
| eva1ba | 0.231 | mdkb | -0.114 |
| pmela | 0.230 | akap12a | -0.113 |
| tspan36 | 0.229 | unm-sa821 | -0.110 |
| dct | 0.226 | hbae3 | -0.109 |
| pim1 | 0.226 | ponzr1 | -0.104 |
| bace2 | 0.226 | s100v2 | -0.104 |
| XLOC-001964 | 0.225 | si:ch73-89b15.3 | -0.102 |
| si:zfos-943e10.1 | 0.224 | prx | -0.100 |
| si:ch73-389b16.1 | 0.222 | ptmaa | -0.099 |
| slc7a5 | 0.222 | krt18b | -0.097 |
| tyrp1a | 0.221 | sytl2b | -0.095 |
| cdkn1ca | 0.218 | slc25a36a | -0.094 |
| tyrp1b | 0.217 | pltp | -0.093 |
| klf6a | 0.215 | si:ch211-105c13.3 | -0.092 |
| slc45a2 | 0.215 | alx4b | -0.091 |
| sypl2b | 0.212 | aqp1a.1 | -0.089 |
| ctsc | 0.212 | prdx5 | -0.088 |
| phlda1 | 0.212 | actc1b | -0.087 |
| pah | 0.212 | gapdhs | -0.087 |
| oca2 | 0.212 | atic | -0.087 |