NLR family member X1
ZFIN 
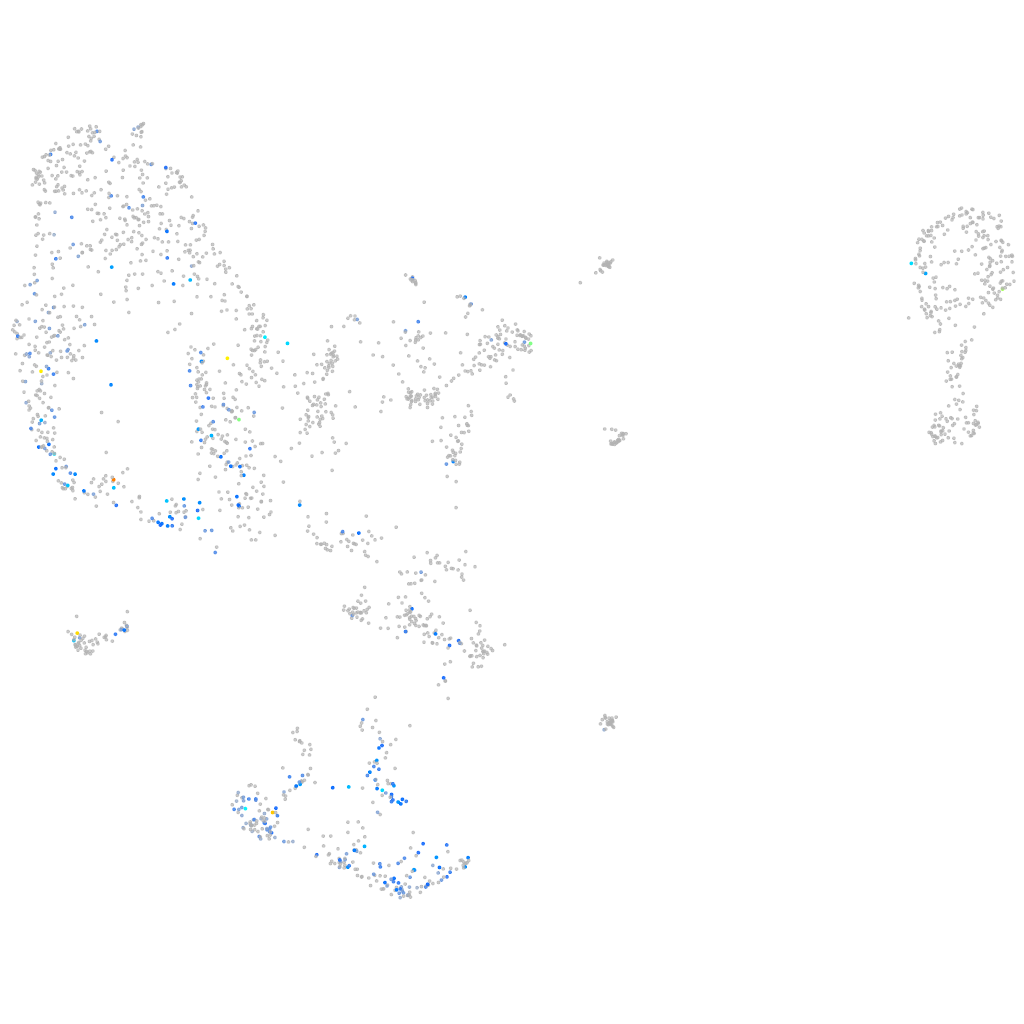
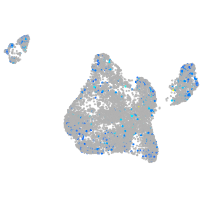
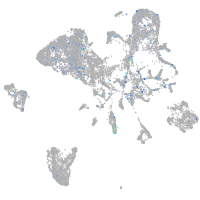
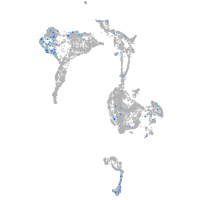
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| malb | 0.246 | hnrnpabb | -0.141 |
| c1qtnf9 | 0.241 | seta | -0.126 |
| bsnd | 0.223 | rpl23a | -0.125 |
| si:dkey-16l2.17 | 0.221 | rpl7 | -0.123 |
| cldnh | 0.221 | rps6 | -0.121 |
| cd9b | 0.209 | marcksl1b | -0.118 |
| chp1 | 0.209 | rplp1 | -0.117 |
| tmem72 | 0.209 | marcksb | -0.116 |
| gdpd3a | 0.206 | apoeb | -0.114 |
| cdh17 | 0.206 | wu:fb97g03 | -0.113 |
| epcam | 0.205 | ebna1bp2 | -0.111 |
| exoc3l2b | 0.204 | hmgb2b | -0.111 |
| sstr5 | 0.201 | si:ch211-51e12.7 | -0.107 |
| si:ch73-359m17.9 | 0.201 | hspb1 | -0.107 |
| si:ch211-207j7.2 | 0.200 | rpl3 | -0.107 |
| tspo | 0.199 | npm1a | -0.107 |
| bin2a | 0.199 | ncl | -0.106 |
| sfxn3 | 0.199 | hnrnpa1a | -0.105 |
| atp1b1a | 0.198 | fbl | -0.104 |
| trim35-12 | 0.198 | cirbpb | -0.104 |
| clcnk | 0.197 | syncrip | -0.104 |
| acsl1b | 0.196 | si:ch211-288g17.3 | -0.103 |
| vdrb | 0.196 | rpl13a | -0.102 |
| LOC101884287 | 0.193 | ran | -0.102 |
| rnf183 | 0.192 | snu13b | -0.102 |
| bcl2l13 | 0.192 | akap12b | -0.101 |
| p3h2 | 0.192 | rps12 | -0.101 |
| me3 | 0.192 | nop58 | -0.101 |
| LOC101887033 | 0.191 | sae1 | -0.100 |
| slc25a48 | 0.191 | setb | -0.100 |
| cdaa | 0.190 | rpl32 | -0.100 |
| ccl19a.1 | 0.189 | rps9 | -0.100 |
| pleca | 0.188 | hnrnpaba | -0.099 |
| tmx2b | 0.188 | nono | -0.099 |
| oclnb | 0.188 | ptges3b | -0.097 |