"neurofilament, medium polypeptide b"
ZFIN 
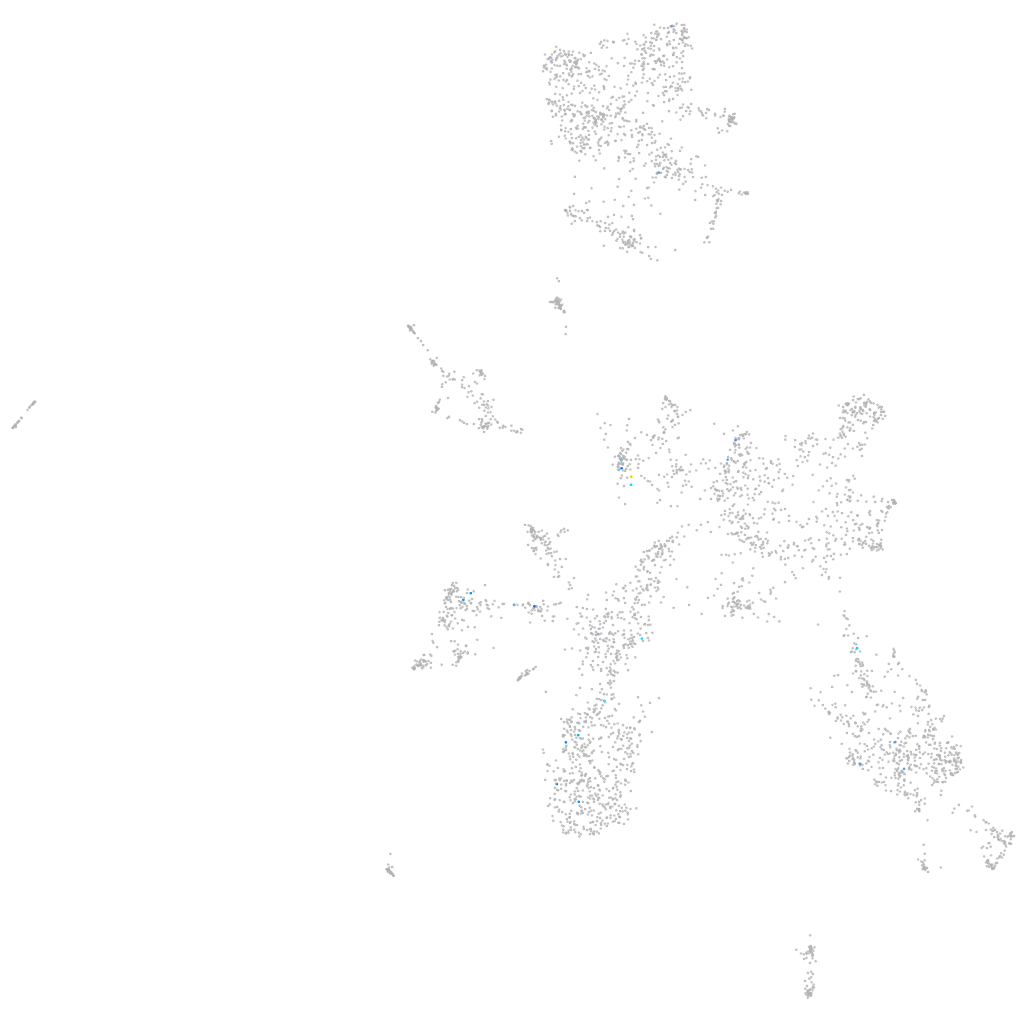
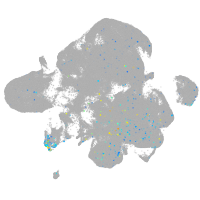
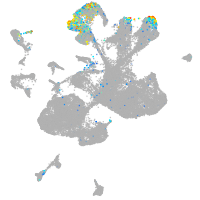
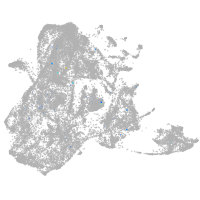
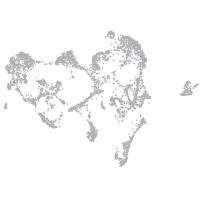
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcdh2ab1 | 0.476 | zgc:56493 | -0.069 |
| hoxc6b | 0.318 | ubb | -0.053 |
| tm6sf2 | 0.299 | prelid3b | -0.050 |
| BX649313.1 | 0.284 | ucp2 | -0.047 |
| BX784029.3 | 0.284 | btg2 | -0.047 |
| prob1 | 0.279 | eef1da | -0.047 |
| XLOC-012620 | 0.273 | GCA | -0.046 |
| he1.2 | 0.265 | pno1 | -0.046 |
| FO704786.1 | 0.263 | psmd14 | -0.042 |
| sall3a | 0.249 | fam53b | -0.041 |
| LOC103910976 | 0.237 | eif4bb | -0.039 |
| sfrp1b | 0.230 | foxp4 | -0.039 |
| XLOC-006573 | 0.214 | si:ch211-286b5.5 | -0.038 |
| CR854989.1 | 0.210 | jun | -0.038 |
| onecut3b | 0.209 | lysmd2 | -0.038 |
| zgc:175280 | 0.201 | eif3ha | -0.038 |
| XLOC-014349 | 0.199 | sod1 | -0.036 |
| XLOC-044229 | 0.196 | reep3b | -0.036 |
| prph | 0.192 | cldnb | -0.036 |
| hoxb6b | 0.192 | epcam | -0.036 |
| socs4 | 0.189 | hif1al | -0.036 |
| gucy1a1 | 0.182 | lin52 | -0.036 |
| itln3 | 0.180 | cebpb | -0.036 |
| XLOC-021043 | 0.180 | rpz5 | -0.035 |
| dlg4b | 0.179 | ddx21 | -0.035 |
| phox2bb | 0.177 | coa3a | -0.035 |
| asic1c | 0.175 | mgst3a | -0.035 |
| gata2b | 0.171 | ccng1 | -0.035 |
| slc18a3a | 0.171 | ahnak | -0.035 |
| RNF122 | 0.170 | cfl1l | -0.035 |
| onecut1 | 0.170 | zgc:92380 | -0.035 |
| CU927934.3 | 0.164 | nr4a1 | -0.035 |
| cacna2d3 | 0.163 | slc25a55a | -0.035 |
| mc5rb | 0.161 | sox2 | -0.035 |
| BX957322.1 | 0.161 | lgals3b | -0.035 |